Modulation of microbial communities in virus infected hosts
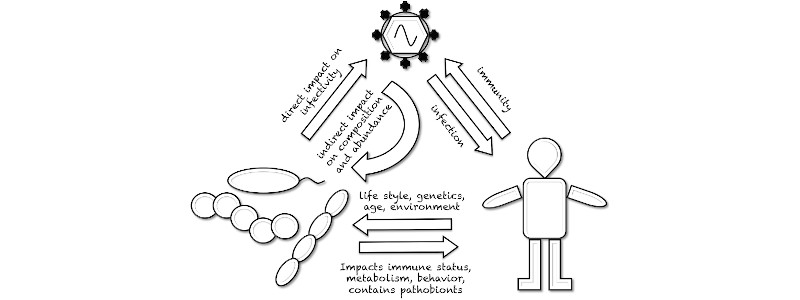
We are living in a densely colonized, poly-microbial environment and even our outer and inner body surfaces are largely covered with microbes. My group in the department of microbiology and molecular medicine studies the impact of acute respiratory infections on the colonization of mucosal surfaces with commensal ("good") and pathological bacteria. We were amongst the first to describe the disturbance of microbial community composition in the context of an influenza A virus (IAV) infection and the role inflammatory processes play in it. Now, we use this knowledge to develop novel antibacterial therapies based on probiotics. We study the function of viral accessory proteins on the innate/inflammatory host immune response on a molecular level. We recently found that a short viral peptide (PB1-F2) is responsible for the antagonism of the NLRP3 inflammasome. This multiprotein complex is responsible for the release of IL1-beta, a key cytokine in proinflammatory responses and important for the host response to viral infections.
Specific expertise
My group has all the genetic tools for manipulation of IAV and we have years of experience with in vitro and in vivo infection models. Using bioinformatic approaches we also characterize the composition of commensal microbial communities on mucosal surfaces under IAV infection conditions.
Selected Publications
Influenza A viruses balance ER stress with host protein synthesis shutoff. Mazel-Sanchez B, Iwaszkiewicz J, Pereira Bonifacio Lopes JP, Silva F, Niu C, Strohmeier S, Eletto D, Krammer F, . Proceedings of the National Academy of Sciences 2021;118(36):e2024681118.
Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Kaul D, Rathnasinghe R, Ferres M, Tan GS, Barrera A, Pickett BE, Methe BA, Das SR, Budnik I, Halpin RA, Wentworth D , Schmolke M, Mena I , Albrecht RA, Singh I, Nelson KE, García-Sastre A, Dupont CL, and Medina RA. Nature Communications 2020;11(1):2537.
Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B, and Schmolke M. Microbiome 2018;6(1):9.
RIG-I detects mRNA of intracellular Salmonella enterica serovar Typhimurium during bacterial infection. Schmolke M, mBio. 2014 Apr 1;5(2):e01006-14
Differential contribution of PB1-F2 to the virulence of highly pathogenic H5N1 influenza A virus in mammalian and avian species. Schmolke M, Manicassamy B, Pena L, Sutton T, Hai R, Varga ZT, Hale BG, Steel J, Pérez DR, and García-Sastre A. PLoS Pathogens. 2011 Aug;7(8):e1002186
24 Mar 2022