Comprehensive catalog of mouse microbiome
Large number of microorganisms live in our digestive tract, from where they play an indispensable role in digestion and influence our health in many ways. To understand how microbes impact the health of the whole body, researchers usually use the mouse as a model. However, the microbial species living in the mouse gut remain poorly characterized.
A new tool to analyze huge quantities of genomic sequences
When Silas Kieser started working in the laboratory of Prof. Mirko Trajkovski with the aim of studying the interaction between the microbiota and its host, the researchers faced with a major problem: the (genome) references of the mouse microbiome were so scarce, that even the latest sequencing-based technologies did not allow identifying and quantifying the species living in laboratory mice.
Therefore, they started by developing a user-friendly tool called metagenome-atlas that allows generating these references from the genomic sequences and thus enabling analysis of any microbiome (more details in their publication in BMC Bioinformatics).
Birth of a catalog of the mouse microbiome
With this new tool in hand, researchers from Pr. Mirko Trajkovski laboratory in tight collaboration with Pr. Evgeny Zdobnov, analyzed all publicly available metagenomes from the mouse gut and established a nearly complete catalog of bacterial species. As described in their recent study published in PLoS Computational Biology, the scientists revealed an incredibly high diversity (see figure below), increasing the percentage of identified species from around 20% to over 90%. They discovered new bacterial families, as well as hundreds of uncultivated species. This enabled the researchers to directly compare the mouse gut microbiome to its human counterpart, highlighting the similarities and dissimilarities between the two.
These results warrant analysis of new datasets and reanalysis of existing ones at an unprecedented depth and can boost research on the microbiota and its influence on health. They also pave the way for new means to implement the microbiota-related findings in mice for human use.
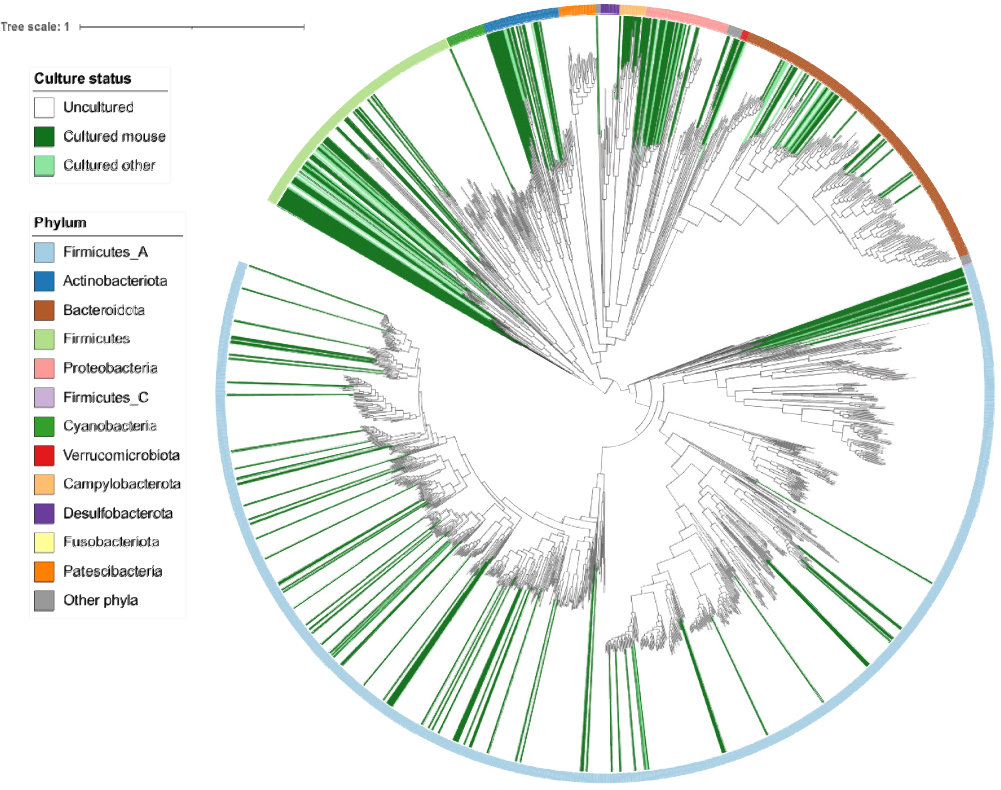
Representation of the bacterial species detected in the mouse gut (each branch in grey corresponds to a species). © from Figure 2 in Kieser et al. 2022 PLoS Computational Biology.
21 Mar 2022
