News
eLife Cell Biology, 2023, 12, RP8865
We suggest that the translational reprogramming called ‘rewiring stress response’ is a protective process of chronic stress adaptation, that regulates cell aging.
Hsf1 and the molecular chaperone Hsp90 support a ‘rewiring stress response’ leading to an adaptive cell size increase in chronic stress
 Summary
Summary
Cells are exposed to a wide variety of internal and external stresses. Although many studies have focused on cellular responses to acute and severe stresses, little is known about how cellular systems adapt to sublethal chronic stresses. Using mammalian cells in culture, we discovered that they adapt to chronic mild stresses of up to two weeks, notably proteotoxic stresses such as heat, by increasing their size and translation, thereby scaling the amount of total protein. These adaptations render them more resilient to persistent and subsequent stresses. We demonstrate that Hsf1, well known for its role in acute stress responses, is required for the cell size increase, and that the molecular chaperone Hsp90 is essential for coupling the cell size increase to augmented translation. We term this translational reprogramming the ‘rewiring stress response’, and propose that this protective process of chronic stress adaptation contributes to the increase in size as cells get older, and that its failure promotes aging.
Cell Reports, 2023, 42, 112786
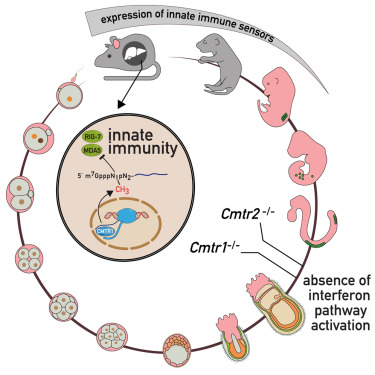
We determined that mammalian cap1 and cap2 modifications have essential roles in gene regulation beyond their role in helping cellular transcripts to evade the innate immune system.
Essential roles of RNA cap-proximal ribose methylation in mammalian embryonic development and fertility
Summary
Eukaryotic RNA pol II transcripts are capped at the 5′ end by the methylated guanosine (m7G) moiety. In higher eukaryotes, CMTR1 and CMTR2 catalyze cap-proximal ribose methylations on the first (cap1) and second (cap2) nucleotides, respectively. These modifications mark RNAs as “self,” blocking the activation of the innate immune response pathway. Here, we show that loss of mouse Cmtr1 or Cmtr2 leads to embryonic lethality, with non-overlapping sets of transcripts being misregulated, but without activation of the interferon pathway. In contrast, Cmtr1 mutant adult mouse livers exhibit chronic activation of the interferon pathway, with multiple interferon-stimulated genes being expressed. Conditional deletion of Cmtr1 in the germline leads to infertility, while global translation is unaffected in the Cmtr1 mutant mouse liver and human cells. Thus, mammalian cap1 and cap2 modifications have essential roles in gene regulation beyond their role in helping cellular transcripts to evade the innate immune system.
Nucleic Acids Research, 2023, 51. 5022
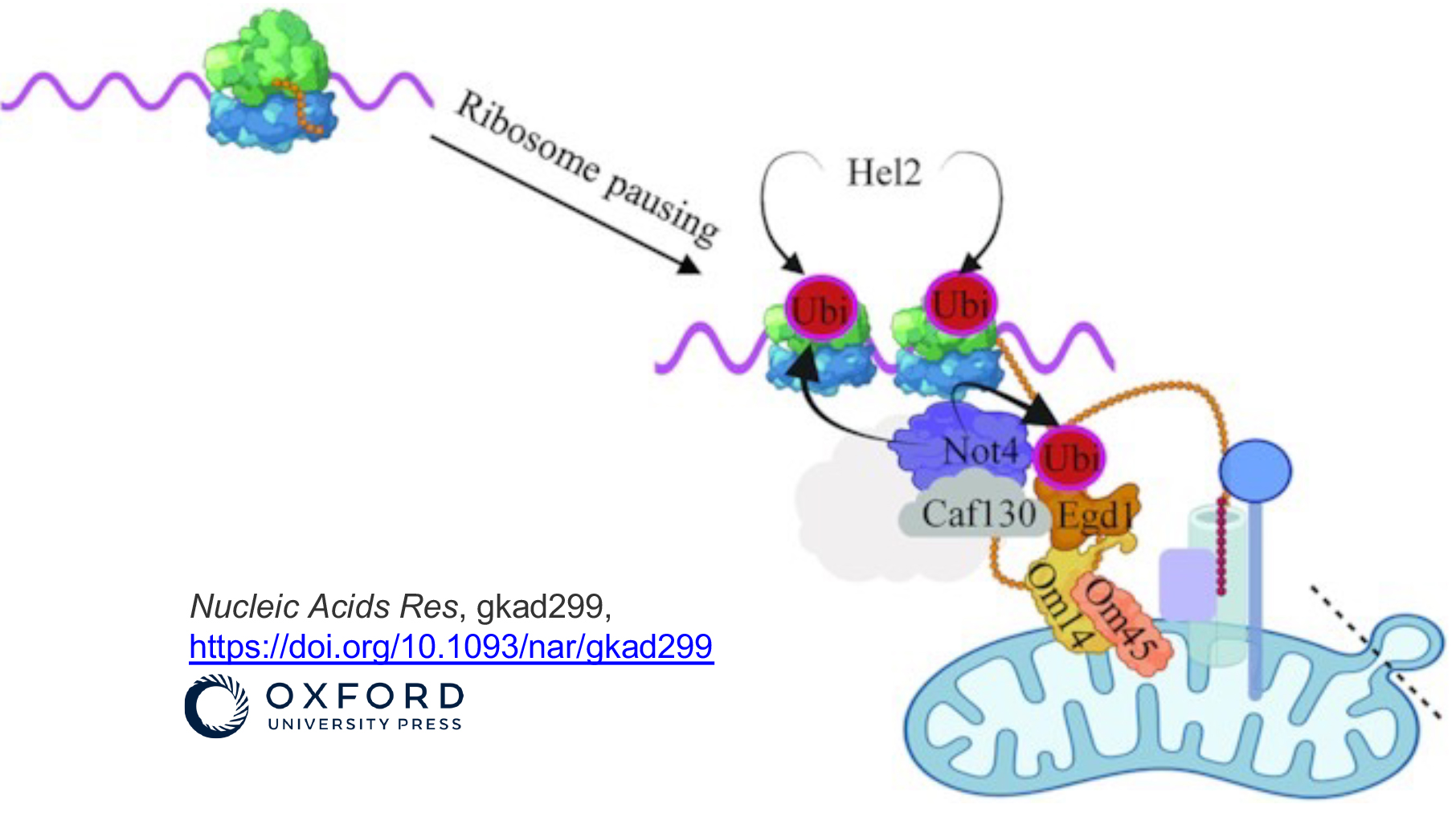
We determined that the Not1 and Not4 subunits of Ccr4-Not inversely regulate mRNA solubility and thereby impact dynamics of co-translation events.
Allen G.E., Panasenko O.O., Collart M.A.
Not4-dependent targeting of MMF1 mRNA to mitochondria limits its expression via ribosome pausing, Egd1 ubiquitination, Caf130, no-go-decay and autophagy
Abstract
The Ccr4–Not complex is a conserved multi protein complex with diverse roles in the mRNA life cycle. Recently we determined that the Not1 and Not4 subunits of Ccr4–Not inversely regulate mRNA solubility and thereby impact dynamics of co-translation events. One mRNA whose solubility is limited by Not4 is MMF1 encoding a mitochondrial matrix protein. In this work we uncover a mechanism that limits MMF1 overexpression and depends upon its co-translational targeting to the mitochondria. We have named this mechanism Mito-ENCay. This mechanism relies on Not4 promoting ribosome pausing during MMF1 translation, and hence the co-translational docking of the MMF1 mRNA to mitochondria via the mitochondrial targeting sequence of the Mmf1 nascent chain, the Egd1 chaperone, the Om14 mitochondrial outer membrane protein and the co-translational import machinery. Besides co-translational Mitochondrial targeting, Mito-ENCay depends upon Egd1 ubiquitination by Not4, the Caf130 subunit of the Ccr4–Not complex, the mitochondrial outer membrane protein Cis1, autophagy and no-go-decay.
Genome Biology, 2023, 24, 30
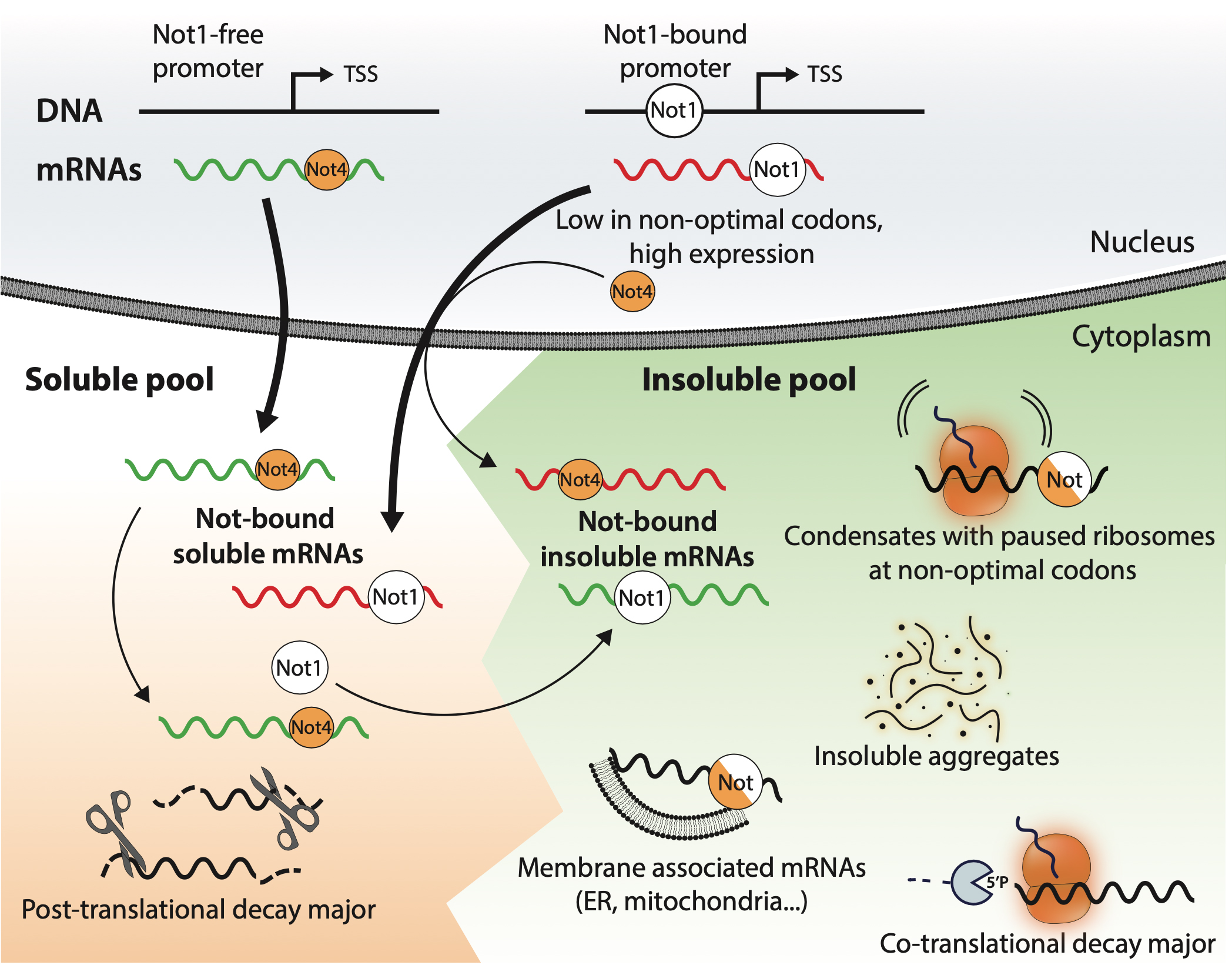
We demonstrated that mRNA solubility defines the dynamics of co-translation events and is oppositely regulated by Not1 and Not4, a mechanism that we additionally determine may already be set by Not1 promoter association in the nucleus.
Allen G.E., Weiss B., Panasenko O.O., Huch S., Villanyi Z., Albert B., Dilg D., Zagatti M., Schaughency P., Liao S.E., Corden J., Polte C., Shore D., Ignatova Z., Pelechano V., Collart M.A.
Not1 and Not4 inversely determine mRNA solubility that sets the dynamics of co-translational events
Summary
In this work, by analyzing soluble and insoluble mRNA decay intermediates in yeast, we determine that insoluble mRNAs are enriched for ribosomes dwelling at non-optimal codons compared to soluble mRNAs. mRNA decay is higher for soluble RNAs, but the proportion of co-translational degradation relative to the overall mRNA decay is higher for insoluble mRNAs. We show that depletion of two subunits of the Ccr4-Not complex, Not1 and Not4, inversely impacts mRNA solubilities and, for soluble mRNAs, ribosome dwelling according to codon optimality. Depletion of Not4 solubilizes mRNAs with lower non-optimal codon content and higher expression that are rendered insoluble by Not1 depletion. By contrast, depletion of Not1 solubilizes mitochondrial mRNAs, which are rendered insoluble upon Not4 depletion.
Activity of R2P Platform was reported at the Swiss RNA Workshop 2023 in Bern


Cell Reports, 2021, 36, 109633
Using the dynamic observation technique, Ribo-Seq, we have deciphered the genetic mechanisms governing the speed of translation of messenger RNA. We show that codon-specific translation elongation dynamics are modulated by Not4 and Not5, in coordination with Rli1 and opposing eIF5A function, and according to codon optimality to produce a soluble proteome. We propose that this regulation occurs by dynamic condensates that limit mRNA solubility and exclude eIF5A.
Allen G.E., Panasenko O.O., Villanyi Z., Zagatti M., Weiss B., Pagliazzo L., Huch S., Polte C., Zahoran Z., Hughes C.S., Pelechano V., Ignatova Z., Collart M.A.
Not4 and Not5 modulate translation elongation by Rps7A ubiquitination, Rli1 moonlighting, and condensates that exclude eIF5A
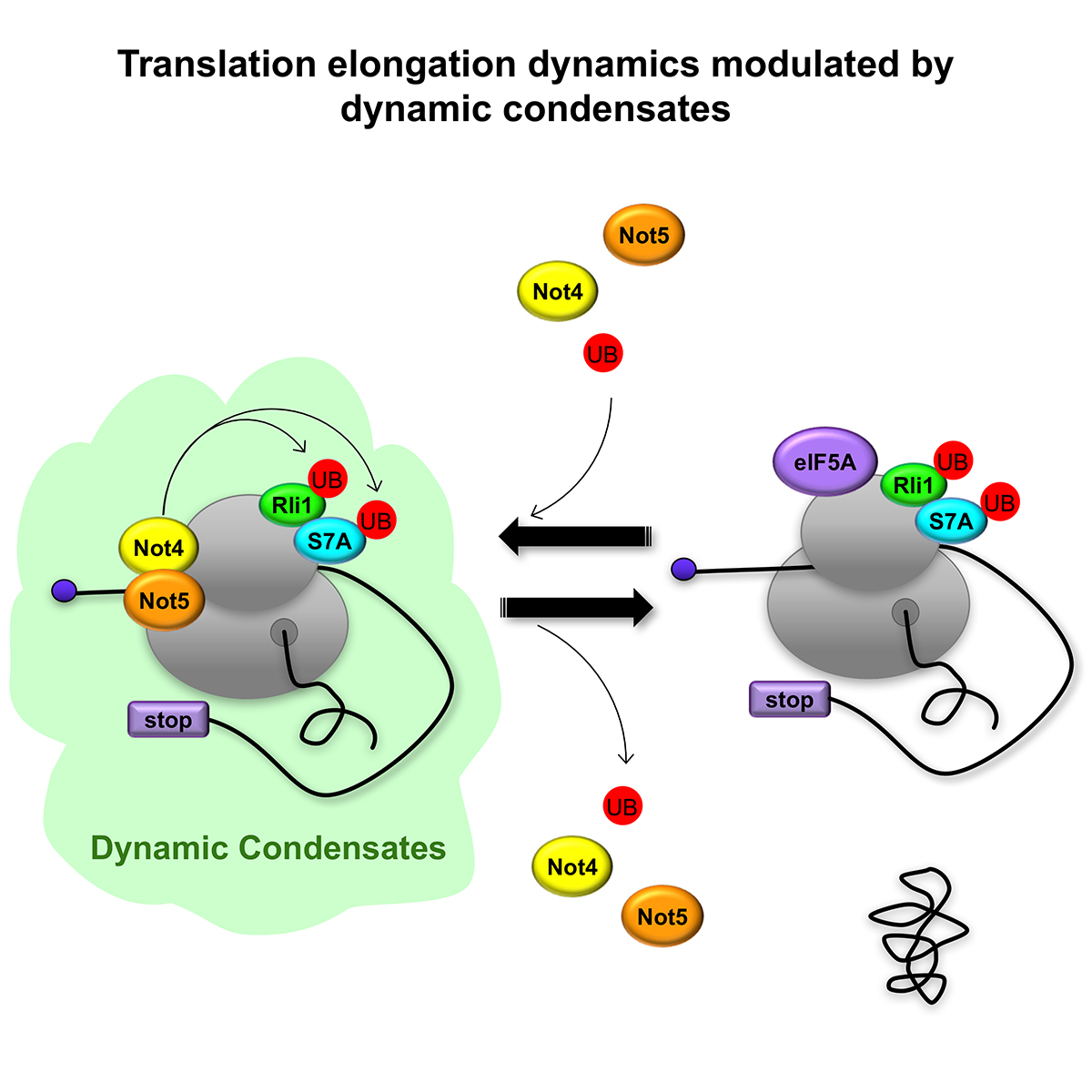
Summary
Nucleic Acids Research, 2021, 49, 2085-2101
Using RNA-Seq, Ribo-Seq and quantitative proteomics we characterized changes in transcriptome, translatome and proteome in pathogen bacteria Staphylococcus aureus.
MazF toxin causes alterations in Staphylococcus aureus transcriptome, translatome and proteome that underlie bacterial dormancy.
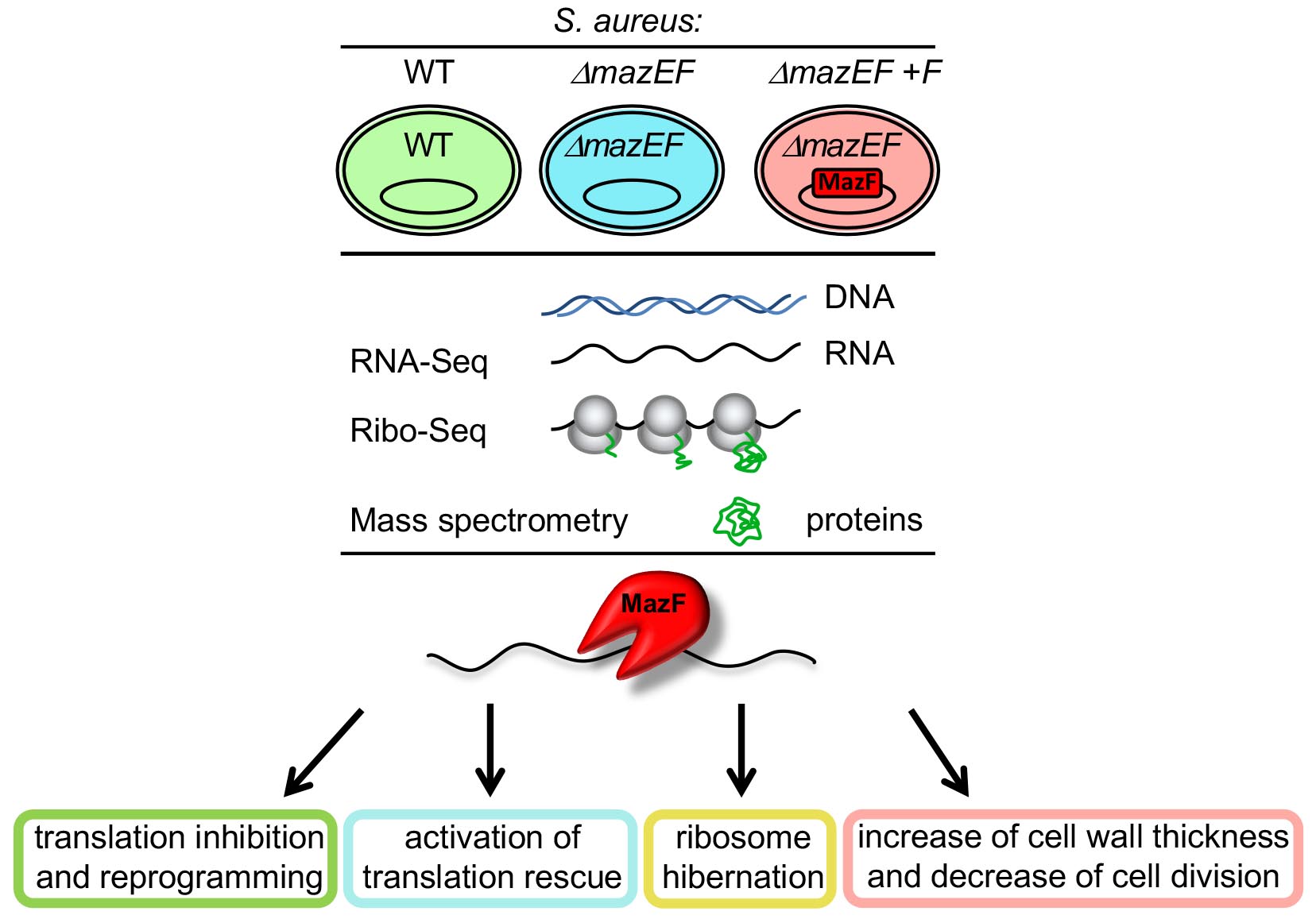
Abstract
Antibiotic resistance is a serious problem which may be caused by bacterial dormancy. It has been suggested that bacterial toxin-antitoxin systems induce dormancy. We analyzed the genome-wide role of Staphylococcus aureus endoribonuclease toxin MazF using RNA-Seq, Ribo-Seq and quantitative proteomics. We characterized changes in transcriptome, translatome and proteome caused by MazF, and proposed that MazF decreases translation directly by cleaving mRNAs, and indirectly, by decreasing translation factors and by promoting ribosome hibernation. Important pathways affected during the early stage of MazF induction were identified: MazF increases cell wall thickness and decreases cell division; MazF activates SsrA-system which rescues stalled ribosomes, appearing as a result of MazF mRNA cleavage. These pathways may be promising targets for new antibacterial drugs that prevent bacteria dormancy. Finally, we described the overall impact of MazF on S. aureus cell physiology, and propose one of the mechanisms by which MazF might regulate cellular changes leading to dormancy.
Cell Reports, 2020, 30, 3851
Using polysome fractionation and Ribo-Seq we demonstrated that peptidyl-prolyl-cis-trans-isomerase (PPIase) FK506-binding protein 10 (FKBP10) interacts with ribosomes, and its downregulation leads to reduction of translation elongation at the beginning of open reading frames (ORFs), particularly upon insertion of proline residues. Our data unveil FKBP10 as a cancer-selective molecule with a key role in translational reprogramming, stem-like traits, and growth of lung cancer.
FKBP10 Regulates Protein Translation to Sustain Lung Cancer Growth
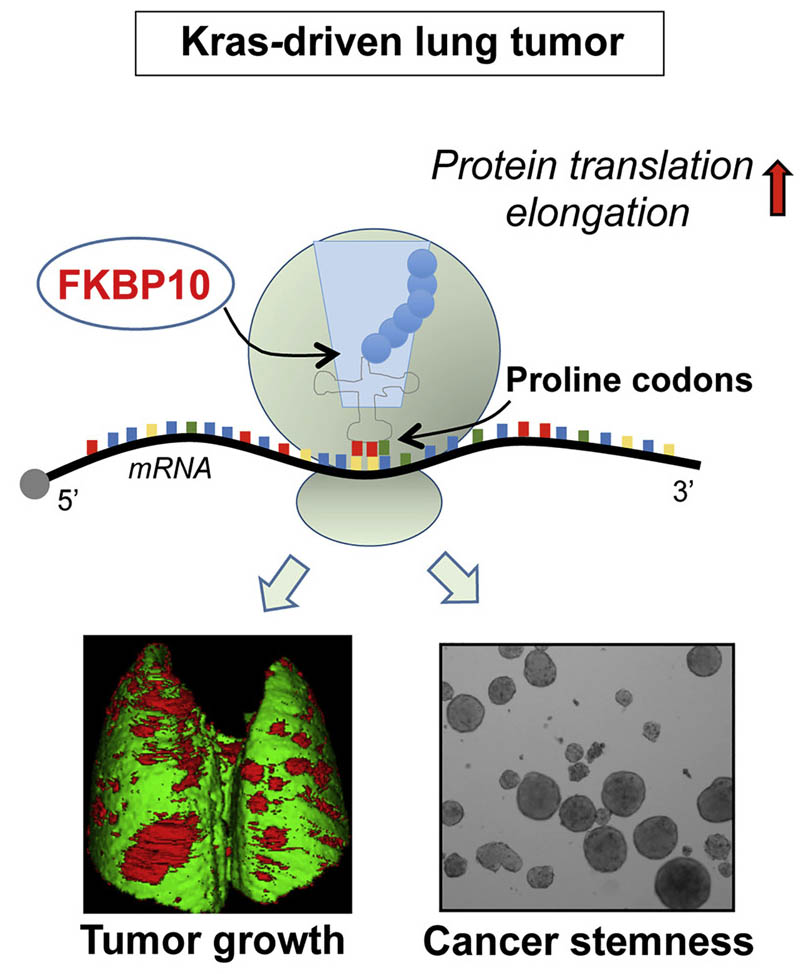
Summary
Cancer therapy is limited, in part, by lack of specificity. Thus, identifying molecules that are selectively expressed by, and relevant for, cancer cells is of paramount medical importance. Here, we show that peptidyl-prolyl-cis-trans-isomerase (PPIase) FK506-binding protein 10 (FKBP10)-positive cells are present in cancer lesions but absent in the healthy parenchyma of human lung. FKBP10 expression negatively correlates with survival of lung cancer patients, and its downregulation causes a dramatic diminution of lung tumor burden in mice. Mechanistically, our results from gain- and loss-of-function assays show that FKBP10 boosts cancer growth and stemness via its PPIase activity. Also, FKBP10 interacts with ribosomes, and its downregulation leads to reduction of translation elongation at the beginning of open reading frames (ORFs), particularly upon insertion of proline residues. Thus, our data unveil FKBP10 as a cancer-selective molecule with a key role in translational reprogramming, stem-like traits, and growth of lung cancer.
Nature Structural & Molecular Biology, 2019, 26, 110
We revealed a mechanism of ribosome pausing and co-translational assembly of Rpt1 and Rpt2, that ensures accurate production of the yeast proteasome.
Panasenko O.O, Somasekharan S.P., Villanyi Z., Zagatti M., Bezrukov F., Rashpa R., Cornut J., Iqbal J., Longis M., Carl S.H., Peña C., Panse V.G., Collart M.A.
Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes
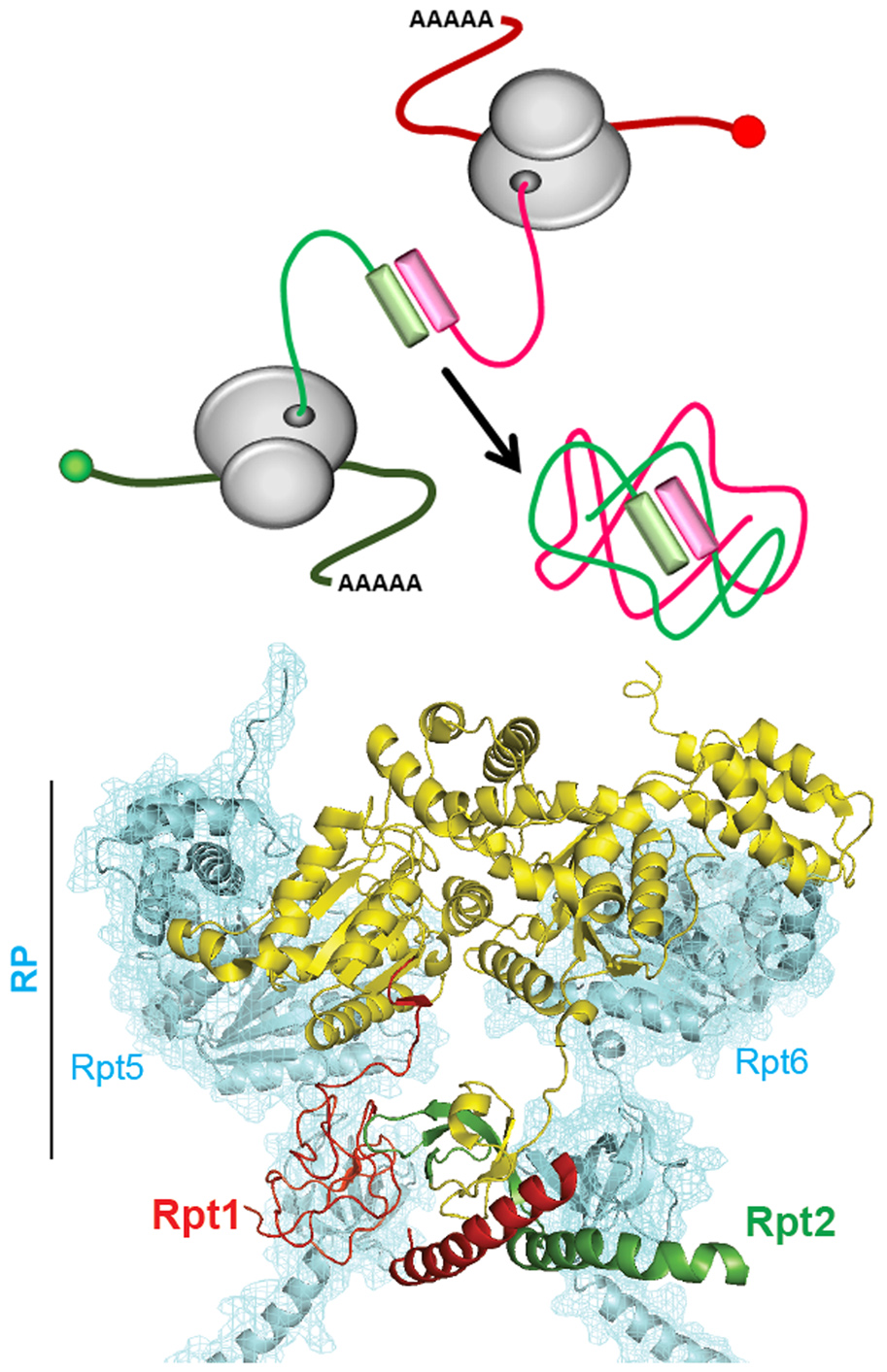
The assembly of large multimeric complexes in the crowded cytoplasm is challenging. Here we reveal a mechanism that ensures accurate production of the yeast proteasome, involving ribosome pausing and co-translational assembly of Rpt1 and Rpt2. Interaction of nascent Rpt1 and Rpt2 then lifts ribosome pausing. We show that the N-terminal disordered domain of Rpt1 is required to ensure efficient ribosome pausing and association of nascent Rpt1 protein complexes into heavy particles, wherein the nascent protein complexes escape ribosome quality control. Immunofluorescence and in situ hybridization studies indicate that Rpt1- and Rpt2-encoding messenger RNAs co-localize in these particles that contain, and are dependent on, Not1, the scaffold of the Ccr4–Not complex. We refer to these particles as Not1-containing assemblysomes, as they are smaller than and distinct from other RNA granules such as stress granules and GW- or P-bodies. Synthesis of Rpt1 with ribosome pausing and Not1-containing assemblysome induction is conserved from yeast to human cells.
