|
What is the Genome Project and why
has it received so much attention?
The genetic information inherited from our parents is contained within the DNA molecules, with about 3.5 billion
bases of DNA in the set of chromosomes received from each parent. This information can be considered to be the
instructions specific for the fabrication of a human being. If it were printed as a large format book with small
print (5000 characters per page) in volumes of 1000 pages, a long row of 700 volumes would be needed. When confronted
with the task of sequencing the entire human genome, scientists were overwhelmed with the task, but in the period
1999-2000 powerful new machines and novel strategies permitted virtually the entire sequence (95%) to be completed
by June, 2000. (The missing sequences are considered to be of only minor significance.) Although this event was
justifiably announced as a major milestone, it is really just the first phase in genome research. In fact, four
distinct stages can be recognized.
What are the different phases of the
Genome Project?
1. Determining the sequence of the entire DNA contents of a human being represents the first stage in the genome
project. This information constitutes the raw data upon which important biological conclusions can be drawn.
2. The second stage, which is now nearing completion, with reports published in SCIENCE and NATURE in special issues
released February 12 , 2001, involves identifying which portions of the DNA sequence correspond to genes. Most
of the DNA sequences do not contain genes; in fact, the roughly 30,000 genes are widely dispersed along the DNA
sequence and occupy less than 2% of the DNA bases). Once the number, size and placement of the genes are established,
many important insights should be forthcoming concerning the numbers of genes in various categories, specifically
how many of the genes are unique and how many are members of gene families with more or less similar structures.
3. The third stage, which is more open ended and will probably last through most of the century, involves understanding
the structure and function of all of the proteins encoded by the genes. Most of the genes identified code for proteins
of unknown function and the long-term challenge will be to devise methods for determining precisely what roles
are played by such proteins.
4. A final stage, for which it is currently difficult to predict exactly what form it will take, involves understanding
the overall structure and function of DNA, in particular what role is played by the non-coding sequences between
the genes and how these sequences contribute to the precise timing and control of expression of the genes that
permits a simple egg to develop into a complex, multi-cellular organism.
What strategies were adopted to sequence
the genome?
In the initial phases of the project doubts were raised as to whether it was feasible to sequence the genome. A
large world-wide consortium was put in place to share the burden and sequencing results were being generated at
a slow, but steady pace, as different specific pieces of chromosomes were assigned to each sequencing laboratory.
However, in the late 1990's a novel approach to DNA sequencing was developed by Craig Venter and his colleagues
in their industrial setting (Celera Genomics) that permitted progress at an accelerating pace. Their method is
based on generating random fragments of DNA to be sequenced, with the final piecing together of the multitude of
sequences achieved by powerful computers. The sequence information obtained by the publicly funded international
consortium had been placed in data bases that were freely available to all scientists without restriction. The
sequence data obtained by Celera is not available without restrictions, so issues of access to the data remain
to be clarified in order to establish how freely available the genome sequence will be for the scientific community.
How difficult was the identification
of genes in the DNA sequence?
Completion of the genome sequence set the stage for the difficult tasks of identifying the portions of the sequence
that correspond to genes and understanding the function of these many, many genes. Basically, the entire DNA sequence
is screened for regions that, when the hypothetical protein sequence encoded by that region is examined, would
correspond to a plausible protein. One way of increasing the confidence that a hypothetical protein is truly a
protein is to find a similar protein in the data bank of protein structures already established (for example, from
other sequenced organisms such as bacteria or yeast). Such criteria, as well as identifying sequences that signal
the beginnings and ends of genes, can help in process. Although a first assessment of the number and location of
the genes can be achieved with current methods, the fine tuning of the identification to improve its reliability
will continue over many years.
Have the goals for sequencing the
human genome changed over the last few years?
The reasons for initiating the sequencing of the human genome were largely for basic knowledge and the somewhat
vague idea that in the long run useful information for human health would be provided. In the last several years,
the motivations turned toward more precise commercial and industrial objectives, as it became obvious that the
financial stakes were high. As portions of the DNA sequence were established and new genes began to be identified,
it became increasingly clear that many were involved in disease states for which the detailed knowledge provided
by the sequence of the gene could help in the design of new drugs and better treatments. Therefore, the rush to
obtain patents and to exploit the findings for drug design and diagnostic kits began, with a key position occupied
by Celera Genomics. As a result the information on the sequence of the genome is being applied rapidly to identify
new genes with medical implications.
What new impacts in human health can
be expected from the Genome Project?
As more and more genes are identified that participate in human illnesses, specific new drugs will be developed
to provide novel therapies for various disease states. Beyond the production of drugs, individualized drug treatments
may be commonplace in the future. Pharmacologists have long recognized that two individuals who have been diagnosed
with the same disease may not respond in the same way to an identical treatment. Individual variations within the
normal range can lead to significant differences in the quantities of proteins in particular tissues. Therefore,
a thorough analysis of an individual in the future will involve specific characterization not only of his or her
genes (genomics), but the level of expression of specific proteins in various tissues (proteomics). This information
will permit treatments of disease states to be highly personalized.
What new approaches in preventive
medicine can be expected?
The identification of the full constellation of genes in a human being and the characterization of the consequence
of mutations in the genes will open new avenues of diagnosis and warnings for dispositions to certain disorders.
Several such tests already exist, as noted for breast cancer, and they may soon be extended to serious personality
disorders, with some indications already appearing for genes involved in schizophrenia. For persons carrying mutant
genes in the heterozygous state (that would be transmitted to a child with a 50% probability, since the child receives
just one chromosome of each pair) opportunities for prenatal diagnosis on an increasingly wide scale will permit
undesirable mutations to be eliminated. The testing can take place on cells obtained at an early stage of a normal
pregnancy, with a therapeutic abortion possible in the event of the identification of a gene that would lead to
a serious disease. Alternatively, several embryos can be generated using the methods of in vitro fertilization,
tested at a pre-implantation stage, and only those embryos without the mutant gene selected for implantation. Such
opportunities involving in vitro fertilization now exist on a limited basis, for certain well-characterized genetic
diseases, in several centers in France and other industrialized countries. For example, for several forms of myopathy,
with a 50% risk to be transmitted to a child if one parent carries the mutant gene, embryos can be screened be
sure that only ones without the mutation are implanted.
What concerns do these developments
raise in terms of ethical considerations and the risks of drifting towards eugenics?
Every form of progress carries intrinsic risks of abuse and the new opportunities in biotechnology can readily
be imagined to be carried to an excess as more and more genes are identified that influence stature, personality,
or intelligence. It is then a short step to suppose that future parents may request prenatal diagnosis to permit
selection of their preferences. Legislative bodies are already alerted and at work in many of the countries where
such initiatives could occur. The issues at stake are widely discussed and touch basic philosophical questions,
as well as purely practical ones. Such testing would require a large infra-structure, if applied on a wide scale,
with costs that would be difficult to justify outside of cases of obvious medical necessity. Questions of public
versus private financing and surveillance by regulatory authorities must also be addressed. While
the scientific community and all concerned citizens must remain vigilant, the nature of the risks at the current
time are still sufficiently vague so that it is premature to attempt to supply detailed responses.
How can genetic information be used
to identify individuals?
Although the DNA sequence of all individuals is extremely similar, a number of minor differences exist throughout
the genome. With each new generation, copying the DNA to make an egg or sperm, although remarkably accurate, is
not perfect and some minor changes in the DNA sequences occur. In the vast majority of instances, these changes
are without consequences, either because they do not make a significant change in genes coding for proteins or
because they occur in regions of the DNA between genes. Because of these minor differences, no two individuals
have exactly the same sequence of DNA down to the last base (except for identical twins). As a result, DNA provides
the potential for uniquely identifying individuals, to a degree and precision that is far superior to any other
method. In practice, there are about a dozen sites on different chromosomes that tend to have the most prevalent
variations between individuals. Hence, by examining these sites, a "DNA fingerprint" can be established
that provides a very high probability of being distinct for every individual examined.
How is DNA fingerprinting carried
out?
Several types of differences in the DNA from one individual to another have been established. The most simple variations
are changes in single bases - in genes as well as in non-coding regions - and these are dispersed throughout the
chromosomes. However, because they occur randomly at all positions, no simple method has been developed to use
single base changes. Another type of variation involves short sequences that repeat a certain number of times with
different numbers of repeats from one individual to another. These sequences, known as STRs (the abbreviation for
Short Tandem Repeats) are particularly well suited to detection by modern methods, since they can be detected as
bands on a gel whose position varies according to the number of repeats present. Tetranucleotide repeats have been
favored for these purposes, typically those with a variation in the human population in the range of 5-15 repeats.
When many different STR regions are examined, the pattern becomes highly specific for each individual.
How reliable are the differences in identity that can be established by DNA?
The recent developments in forensic DNA are based on examination of 13 STRs on different chromosomes. The different
combinations are readily visualized on "ladders" of differently spaced bands that provide a specific
pattern of bands, with a sufficiently large number combinations that the chances of two randomly selected, unrelated
individuals having the same pattern is one in a trillion. Even for siblings there is on average only a chance of
one in 40,000 for the same profile to appear. These statistics make DNA testing far more reliable for identification
of individuals than blood groups or other measurements based on proteins.
What are the current applications of DNA testing?
The principal impact of DNA testing has been for crimes. Because so little DNA is required, traces of blood or
semen can provide a "DNA fingerprint" of a criminal that can be used to eliminate or implicate suspects.
Much interest in this method was provided by the 1995 trial of O. J. Simpson, a legendary football star in the
US accused of murdering his wife. Although the DNA evidence appeared to be convincing, the jury found Simpson not
guilty, largely because of racial discrimination by the police. Other applications include questions of paternity
(even involving deceased persons, as in the case of alleged paternity involving Yves Montand) or the identity of
human remains. Similar methods, but involving variations among individuals due to difference in DNA sequence exclusively
on the Y chromosome, were used in 1998 to suggest that Thomas Jefferson had fathered a son with his slave, Sally
Hemings, on the basis of comparisons between descendants of Jefferson and Hemings' son, Thomas Woodson. Sequences
on mitochondrial DNA can also be used for identity, as applied in 1996 to identify the remains of Russian Tsar
Nicholas II among the bodies from a mass grave near Yekaterinburg, Russia.
Are any concerns raised by the application of these methods?
DNA testing has raised concerns in the realm of privacy and human rights. In certain cases of rape and murder where
DNA samples of the alleged perpetrator where available, police have obtained DNA samples from a wide circle of
individuals living or working in the region of the crime. The question is therefore raised as to whether governmental
law enforcement agencies have the right to force individuals to cooperate in providing cells for DNA typing (for
example, by scraping a cotton swab along the lining of the mouth) and most rely on individuals volunteering their
DNA. On a wider scale, armies (notably in the United States) have begun stocking DNA fingerprints of all of their
members to facilitate identification of remains and the possibility of collecting DNA from all individuals of a
country at birth has been raised. These measures have been advocated in several States in the USA and are also
supported by groups that working to combat child-trafficking. Although DNA identities might help to prevent some
abuses, circumstances could be imagined in which an authoritarian government would use such information to reinforce
a police state. In conclusion, the full impact of DNA fingerprinting has not yet been established
and the issue of whether testing on a large scale infringes upon or protects individual rights remains to be settled.
V. UNDERSTANDING LIFE
What can genetics tell us about the
origin of species?
Genetics provides the context in which evolution can be understood. As enunciated by Darwin, evolution depends
on spontaneous variation and natural selection. Although unclear at the time that Darwin published the Origin of
Species (1859), we can now understand how variation arises because of the insights provided by genetics. The key
elements are mutations -- changes in the chromosomes that are "permanent" and can be passed on from one
generation to the next. Mutations arise spontaneously, because "errors" are introduced when chromosomes
duplicate. In the vast majority of cases mutations are harmful, but occasionally a change will be produced that
leads to new properties that endow the organism with a selective advantage in its environment. The advantage will
be translated into a higher rate of reproduction and after many generations, the organism with the new properties
will predominate. When the changes involve a number of major characteristics, the organisms with these characteristics
will have diverged significantly from their recent ancestral forms, creating a new species.
What has gene sequencing taught us
about evolution?
Knowledge about the precise structure of genes has revolutionized studies of evolution. One of the major surprises
for biologists, as the sequences of more and more genes became available, was just how similar the genes can be
from two organisms that would appear to have virtually nothing in common. For example, when human beings and yeast
are considered, there is no outward reason to expect similarities, but the cells of both undergo chromosome duplication
(mitosis) during cell division. It was found that one of the key proteins in this process is so similar in yeast
and human cells that the yeast gene coding for this protein can be successfully replaced by the corresponding human
gene. Overall, in the last several years, gene sequencing has revealed that all living forms share more common
genetic features than had been previously suspected. Indeed, nearly all genes in humans occur in very similar forms
in mice. Yet, mice and humans are clearly very different in many important ways. The reasons are less in the genes
themselves than in the regions of chromosomes between the genes that control the expression of genes. Therefore,
with similar genes that are expressed at different times and in different locations of the body, a fertilized egg
in one case produces a mouse and in the other case a human. The way in which the information between genes leads
to such differences is only beginning to be understood in very preliminary terms.
Can the link between genes and evolution
be explained in detail?
The theory of evolution provides a satisfactory explanation for the diversity of living forms and the appearance
and disappearance of individual species in the fossil record. However, evolution is an extremely complex process
that involves interactions between species (predators and prey among animals; interaction between plants and animals,
with animals both consuming plants and aiding in their dissemination), as well as climatic and geological changes.
For all of these reasons, it has not been possible to provide a precise explanation of the link between genes and
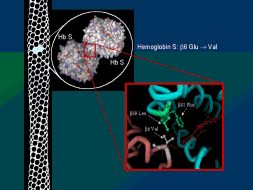
evolution, except in some "model" systems, such as the rise of the sickle cell hemoglobin mutation in
regions where human populations were subject to malaria. However, such "model" systems are more appropriately
designated as "micro-evolution," rather than full evolution, since they involve limited changes within
an existing species, not formation of a new species.
How can the formation of a new character
be explained by evolution?
For the same reasons that evolution of new species cannot be explained in detail, the formation of a major new
character is also a process that can only be imagined in general terms. Indeed, one of the criticisms leveled against
Darwinian evolution by creationists, is precisely the difficulty in explaining how a complex organ such as an eye
or a heart arose, when so many individual steps are required to develop the organ. Since the organ is only fully
functional in its completed form, it is unclear how each individual step was selected because of some advantage,
with the accumulation of the individual steps eventually leading to the fully developed organ. Nevertheless, in
some cases we can begin to formulate plausible hypotheses, for example, concerning the eye. The basic step in vision
is the capture of a photon that induces a change in the protein rhodopsin. We now know that rhodopsin is in the
same family of proteins as the receptors responsible for the recognition of odors. We can therefore imagine an
early organism without vision that happened to have a mutation in one of its odor receptors that allowed it to
bind a light-sensitive molecule. Obviously many additional steps must be imagined to generate an eye and such scenarios
are totally hypothetical. Nevertheless, because we cannot necessarily imagine all of the
steps in a complex process, that is not a reason to embrace creationism -- which would indeed face a number of
insurmountable problems of its own, for example in providing an explanation of how the creator put together all
of the different genomes of all of the species on Earth!
Are genes known in which mutations
lead to a specific change in behavior?
In general, all forms of human behavior are extremely complex and involve the contributions of numerous genes and
environmental influences. For most major psychiatric disorders, such as schizophrenia and depression, tendencies
for these conditions to run in families have been noted, but genetic factors alone are not sufficient (even an
identical twin has only about a 50% change of being affected if the other twin has the disorder) and studies of
genetics in families afflicted suggest that several distinct genes are involved. Nevertheless, in some cases it
has been possible to establish a strikingly clear relationship between a mutant gene and a particular form of behavior.
A classical illustration is the gene for the protein MOA-A (monoamine oxidase A) in which certain mutations produce
aberrant personalities characterized by highly aggressive behavior (random violence, rape, exhibitionism, arson).
The gene that codes for this protein is located on the X chromosome and in the families concerned, only males are
affected, since in females the second X chromosome compensates, as in the case of hemophilia. The cause of the
aggressive behavior is presumably related to a defect in the metabolism of an important brain neuro-transmitter,
but the exact details not understood.
Can the relationship between genes
and behavior be understood using animals?
Many important insights have been obtained using animal models for behavior. Among the most spectacular recent
discoveries concerns narcolepsy, a debilitating sleep disorder (involving excessive daytime sleepiness, including
cataplexy) that affects 1 in 2000 individuals. Little insight into the origins of this condition were available
until investigations were completed on a colony of narcoleptic dogs studied at Stanford University. Extensive genetic
analysis permitted the gene responsible for this condition to be identified, a gene that encodes the receptor for
a neuropeptide known as hypocretin (or orexin). When this peptide was eliminated in mice using the gene knockout
methods, the mice also displayed symptoms of narcolepsy. Based on these observations, studies were conducted on
human patients with narcolepsy and indeed most patients were deficient in hypocretin/orexin. The discovery of the
biochemical basis of narcolepsy has not yet led to a treatment, but these findings represent a promising starting
point.
What other direct links have been
established between genes and behavior?
Striking observations have been made in mice based on genetic changes that can be readily achieved in these animals.
For example, in studies reported by Larry J. Young and his colleagues reported in the August 19, 1999 issue of
Nature, a gene from a highly social, monogamous relative of the mouse, the prairie vole, was transferred to laboratory
mice. The gene encodes for the receptor of the hormone arginine vasopressin and the form present in the prairie
vole includes an adjacent non-coding promoter region that triggers expression in portions of the brain suspected
to be involved in pairbonding. When the mice with the prairie vole form of the gene were injected with arginine
vasopressin, a dramatic increase was observed in the affiliative behavior of males as measured by the time spent
in olfactory investigation and grooming of females. Although many genes may be involved in a complex social behavior
such as monogamy, these results indicate that changes resulting from the expression of a single gene in critical
areas of the brain can have a marked influence as aspects of such behavior, such as affiliation.
Do genetic results shed light on the
issue of innate versus acquired characteristics?
The classical problem of nature versus nurture is not likely to be resolved in a simple way by genetics. For a
complex issue such as human intelligence, it would be difficult to design experiments to distinguish between genetic
and environmental influences on learning, but such experiments can already be carried out in mice, with particularly
interesting results reported by Ya-Ping Tang and his colleagues in the September 2, 1999 issue of Nature. They
worked with receptors for one of the major neurotransmitters in the brain, glutamate. This neurotransmitter is
involved in triggering the passage of signals at synapses between neurons. Many different forms of glutamate receptors
are found in the brain and several of the forms that have been implicated in learning and memory also bind the
compound N-methyl-D-aspartate (NMDA). These so-called NMDA receptors have been implicated in learning and memory,
especially in a region in the center of the brain known as the hippocampus. Moreover, the NMDA receptors exist
in two versions, one that predominates in younger animals, another that predominates in adults. The juvenile form
responds more strongly to glutamate than the adult form. Tang and his coworkers engineered mice to express the
juvenile receptors in large quantities, particularly in the hippocampus. The mice were then subjected to a number
of learning tests: associating a tone with a mild electric shock in a particular environment, recognizing that
a "new" object and spending more time exploring it, and recalling where a submerged platform is located
in a pool of opaque water. In all tests the transgenic mice scored significantly higher than otherwise identical
mice in which the juvenile NMDA receptor gene had not been added. Although our understanding
of the molecular basis of memory is still incomplete, the fact that critical genes may play a role in the "innate"
component of learning is becoming increasingly clear.
Can individual genes be studied for
their impact on longevity?
Aging is a feature of biology that varies strikingly among different species. Consider among mammals for example
the difference between mice and humans. The average life-time for normal laboratory mice in a recent aging study
was 761 days. For humans, in countries with high health standards an average life time is about 75 years. Therefore,
for two species that are composed of similar types of cells and roughly equivalent amounts of DNA per cell and
numbers of genes (most of which produce proteins with very similar properties), the lifetimes differ by a factor
of 36. Most probably important differences in a number of critical genes are responsible for this factor of 36,
but identifying which genes are involved is a major challenge. One approach would be to study the genomes of individuals
who live to exceptionally old ages (particularly if family studies indicate an inherited tendency to longevity)
to determine whether particular properties can be identified. Studies along these lines may benefit from the availability
of island populations with excellent genealogical records, such as found in Iceland. A more experimental approach
is also possible using laboratory animals such as mice, for which selected genes can be modified to determine if
they have an effect on average life times.
In what ways do genes control aging?
How aging is controlled by genes is not yet well understood, since many different genes are probably involved,
making it difficult to establish a distinct effect for a particular gene. Nevertheless, some success has been achieved
in the context of the hypothesis that aging is associated with oxidative damage or UV damage, the most common forms
of environmental stress. In laboratory experiments, exposure to UV light is readily achieved and oxidative damage
can be simulated by treatment with hydrogen peroxide (H2O2). In this context, Enrica Migliaccio and her colleagues
described mice in the November 18, 1999 issue of Nature that had been genetically modified to improve the response
of natural cellular mechanisms that fight the harmful effects of environmental stress. Indeed, under standard laboratory
conditions these mice live longer (an average of 973 days) than the otherwise identical mice without the genetic
modification (an average of the 761 days). These studies represent an important first step towards establishing
experimental models, particularly for mammals, in which hypotheses for genes that control aging can be tested.
Are cells genetically programmed to
age?
All living cells have intrinsic aging properties that can be examined from two perspectives -- the number of times
they divide and the total life time of an individual cell. Concerning division, the category of cells known as
stem cells (i.e., cells capable of differentiating into other specific cell types) possess the particular property
of dividing an unlimited number of times, but most other types of cells, known as somatic cells (i.e., cells fully
differentiated such as muscles or neurons), will divide only a limited number of times. Tumor cells, like stem
cells, are "immortal" can be grown indefinitely in cell culture. Indeed, one of the concerns for using
human stem cells in therapeutic approaches to disease states is that the stem cells may not be adequately controlled
by the new host and may proliferate excessively to create unwanted masses of cells that resemble tumors. With respect
to the time a mature cell lives, even in the same organism, different types of cells have very different life expectancies:
for a red blood cell, total life time is a matter of a few months, whereas certain neurons can live for the entire
lifetime of human being. Nevertheless, as they age human beings do lose a significant percentage of their neurons,
so the average life expectancy of a neuron is less than the 70-80 years of a human being. Moreover, many "extra"
neurons die in the early years of post-natal development. For both red blood cells and neurons, as well as all
other types of cells, specific biochemically-based aging programs are present, but no specific information is yet
available to establish precisely how cell lifetimes are determined.
Is aging determined by a single gene
or by multiple genes?
Although a limited number of genes have been identified that can influence aging in a particular environment, the
overall process of aging and the factors responsible for species-dependant life expectancies are certainly under
the control of many different genes. We note that all children generally pass through very similar stages of development
at similar ages, but the changes of old age are much more variable and affect different individuals in different
ways. One reason may be that aging, as it applies to "seniors," generally occurs after individuals have
completed their reproductive phase. As a result, aging may have largely escaped from the evolutionary process of
natural selection, and the factors involved in aging may be less homogeneous than the factors involved in development
from conception to maturation at the young adult stage. Therefore, it is unlikely that one or a small number of
critical "aging" genes will be identified. Alas, the utopian dream of achieving immortality is not about
to be aided by progress in genetics -- even were it considered to be a goal worth pursing. Perhaps Julien Gracq
had the deepest insight into this matter when he wrote "Notre idée de l'immortalité, ce n'est
guère que la permission pour quelques-uns de continuer à vieillir un peu une fois morts." (Our
idea of immortality is nothing but the permission for some of us to keep aging a little after having died.)
|