News
FebruarY 2024
A new visitor in the team for a few months! 
Carolina Panosso Schuindt, a bachelor student from the Freire Neto group in São Paulo, joined us for three months, thanks to the support of FAPESP, in order to work on the interaction between D. discoideum and a colorful opportunistic pathogen, Chromobacterium violaceum.
"As part of my research at the University of São Paulo (Brazil), I characterized the role of ZntA and ZntR in the context of zinc and cadmium intoxication in the opportunistic pathogen Chromobacterium violaceum(Cv). Now I have joined the Soldati lab for a 3-month internship, and under the supervision of Dr. Lucas Ceseti, I will investigate what happens when Dicty meets Cv and whether ZntA and ZntR are essential for the bacterial virulence in the context of infection." - Carolina Panosso Schuindt
Congratulations to Crisalida for her successful master's defense !
After a year with us, Crisalida successfully defended her master's work. She worked at investigating the role of lipophagy in lipid droplet dynamics in D. discoideum. She will continue her work with us for few months, before to start her PhD in another team at Unige 
January 2024
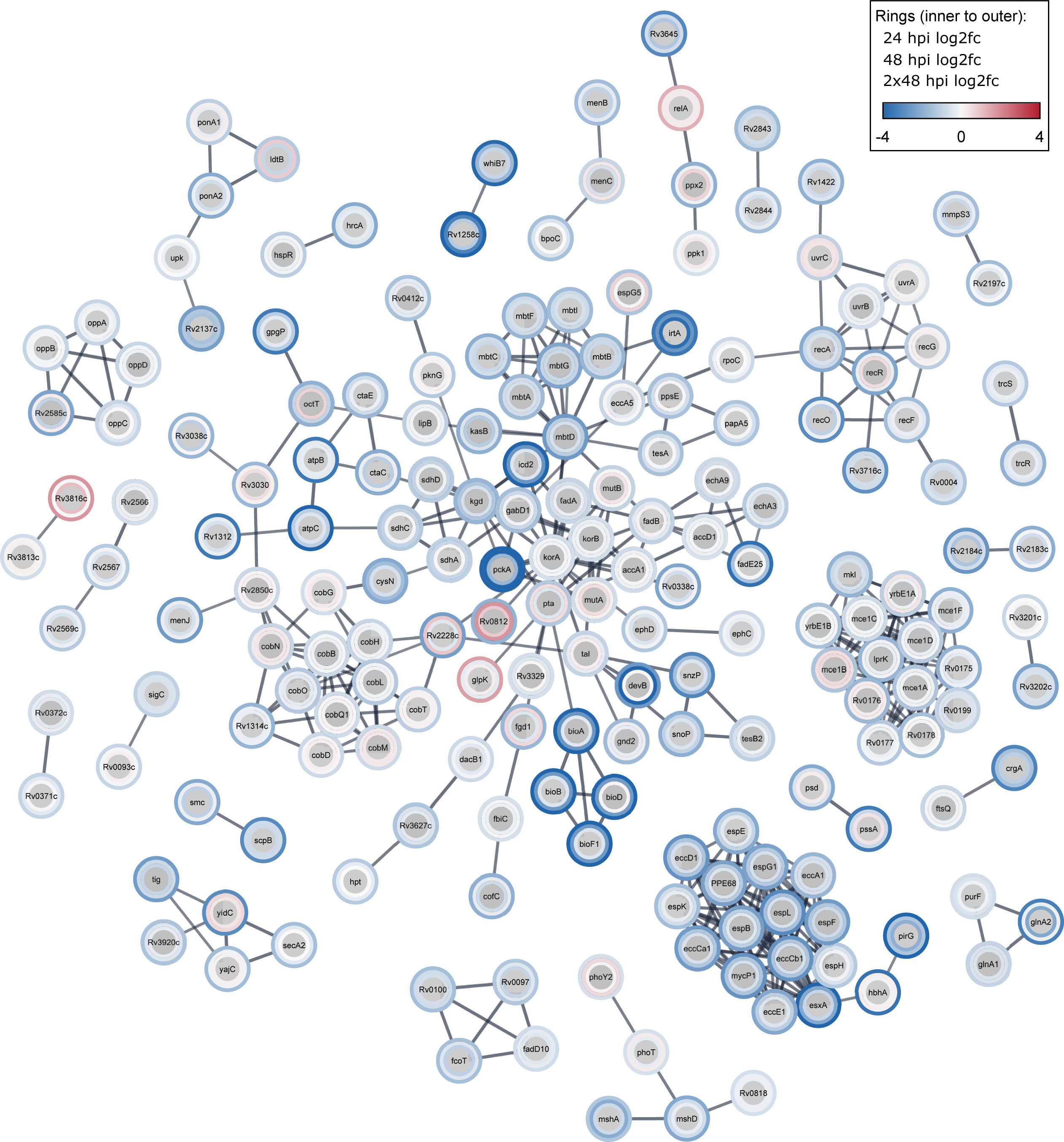
New paper out in mSystems: Temporal genome-wide fitness analysis of Mycobacterium marinum during infection reveals the genetic requirement of virulence and survival in amoebae and microglial cells
In this pivotal study, our lab, in collaboration with the team of Pr. Graham Stewart, has identified critical genes essential for mycobacterial survival during the infection. Tuberculosis, a persistent infectious disease, has become even more challenging with the rise of drug-resistant strains, necessitating improved treatment strategies. Utilizing transposon sequencing (Tn-Seq) on Mycobacterium marinum (a model pathogen for M. tuberculosis), we pinpointed genes crucial for infection survival. This unbiased genome-wide approach highlighted disruptions in 57% of TA sites and identified 568 essential genes (10.2%). Noteworthy pathways for M. marinum survival during infection in different host cells included DNA damage repair, lipid and vitamin metabolism, the type VII secretion system (T7SS) ESX-1, and the Mce1 lipid transport system. These findings, consistent with previous studies, provide valuable insights into tuberculosis pathogenesis, offering potential targets for more effective drug interventions.
For the complete story, click here!
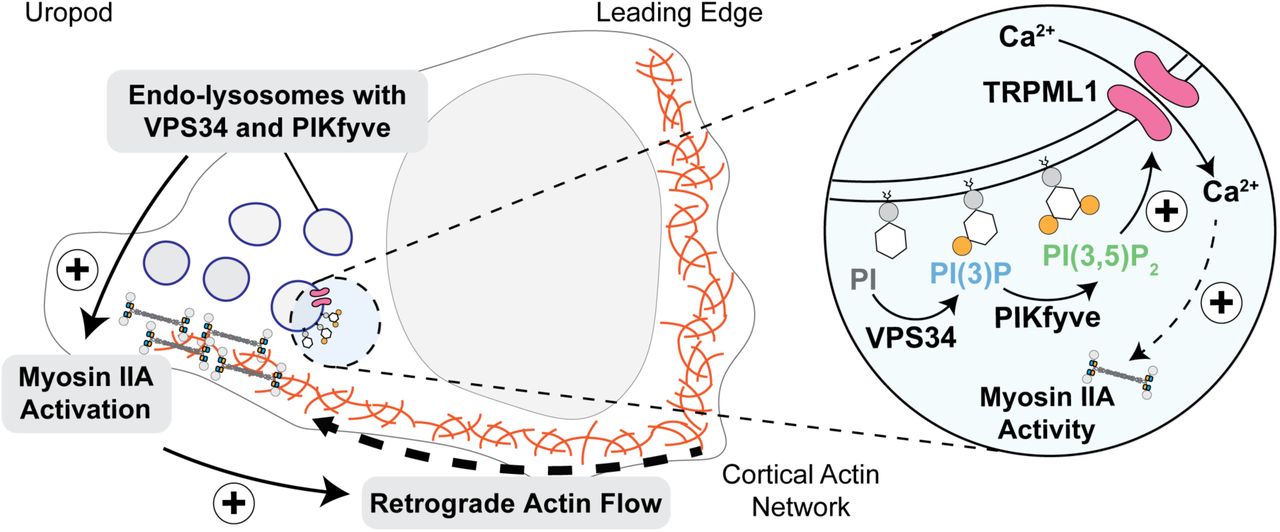
New collaborative paper with Philippe Dehio in the team of Prof. Christoph Hess: An evolutionary-conserved VPS34-PIKfyve-TRPML1-Myosin II axis regulates the speed of amoeboid cell migration
In this new collaborative work, a crucial connection between ultrastructural polarization and T cell migration signaling have been unveiled. This study led by the team of Prof. Hess, identifies endo-lysosome-localized kinases, specifically VPS34–PIKfyve, as instrumental in regulating the speed of amoeboid migration in T cells. The research demonstrates the accumulation of these kinases at the uropod of polarized cells, influencing T cell velocity without altering directionality. The mechanism involves the generation of PI(3,5)P2, controlling Ca2+ efflux through TRPML1 and subsequently regulating myosin IIA activity and propulsive force via retrograde actin flow. Notably, the study also highlights the evolutionary conservation of the VPS34–PIKfyve axis in controlling migration speed in myeloid cells and Dictyostelium discoideum.
Have a look here!
OCTOBER 2023
An SNSF grant has been assigned to our group !
We are delighted to announce that our group has been granted an SNSF grant to work on the virulence strategies of pathogenic mycobacteria and cell-autonomous host defence mechanisms. We are happy to continue our strong contribution to the field with our unique model !

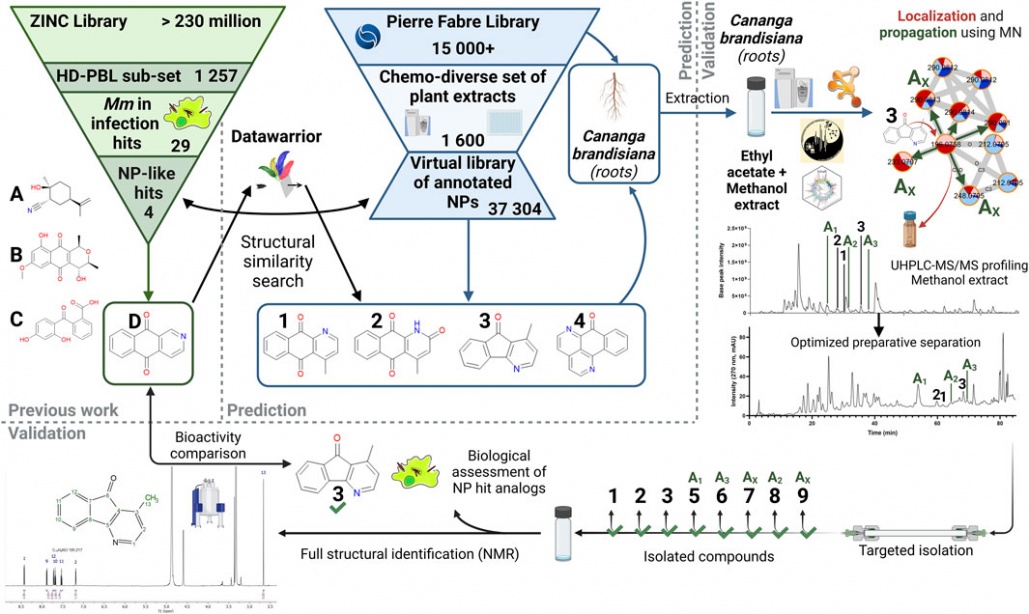
New collaborative paper with the Wolfender group out in Frontiers in Natural Products: Targeted isolation of natural analogs of anti-mycobacterial hit compounds based on the metabolite profiling of a large collection of plant extracts
Antibiotics resistance is a clear threat to the future of current tuberculosis treatments like rifampicin, prompting the need for new treatment options in this field. While plants can offer a plethora of chemical diversity in their constitutive natural products to tackle this issue, finding potentially bioactive compounds in them has not always proven to be that simple. Classical bioactivity-guided fractionation approaches are still trendy, but they bear significant shortfalls, like their time-consuming nature as well as the ever-increasing risk of isolating known bioactive compounds. In this regard, we have developed an alternative method to the latter approach that allows for natural derivatives of a known bioactive scaffold to be efficiently targeted and isolated within a large library of plant extracts. Hence our approach allows for the anticipation of bioactive structure independently of preliminary bioassays. By relying on the chemical diversity of a set of 1,600 plant extracts analyzed by HRMS/MS, we were able to isolate and characterize several minor derivatives of a previously reported bioactive aza-anthraquinone compound from Cananga brandisiana, selected within the plant set. Assessment of bioactivity on these derivatives confirmed their expected activity on Mycobacterium marinum in our anti-infective assay. This proof-of-concept study has established an original path towards bioactive compounds isolation, with the advantage of potentially highlighting minor bioactive compounds, whose activity may not even be detectable at the extract level.
For a complete reading, it's here.
AUGUST 2023
 Master internship
Master internship
We are looking for two motivated master students to join our lab and collaborate on our most recent stories: TrafE and Vacuolins
Don't hesitate to check our Job offers page !
June 2023

Very exciting news! Our postdoc Sandra Guallar-Garrido has been awarded an SNF postdoctoral fellowship!
This fellowship will allow here to keep deciphering the role of TrafE during M. marinum infection in Dicty. Huge congratulations to her!
New collaborative paper with the team of Jason King out in Journal of Cell Biology: A PI(3,5)P2 reporter reveals PIKfyve activity and dynamics on macropinosomes and phagosomes
Phosphoinositide signaling lipids (PIPs) are key regulators of membrane identity and trafficking. Of these, PI(3,5)P2 is one of the least well understood, despite key roles in many endocytic pathways including phagocytosis and macropinocytosis. PI(3,5)P2 is generated by the phosphoinositide 5-kinase PIKfyve, which is critical for phagosomal digestion and antimicrobial activity. However PI(3,5)P2 dynamics and regulation remain unclear due to lack of reliable reporters. Using the amoeba Dictyostelium discoideum we identify SnxA as a highly-selective PI(3,5)P2 -binding protein and characterise its use as a reporter for PI(3,5)P2 in both Dictyostelium and mammalian cells. Using GFP-SnxA we demonstrate that Dictyostelium phagosomes and macropinosomes accumulate PI(3,5)P2 three minutes after engulfment but is then retained differently, indicating pathway-specific regulation. We further find that PIKfyve recruitment and activity are separable, and that PIKfyve activation stimulates its own dissociation. SnxA is therefore a new tool for reporting PI(3,5)P2 in live cells that reveals key mechanistic details of the role and regulation of PIKfyve/PI(3,5)P2 .
Have a look here!
APRIL 2023
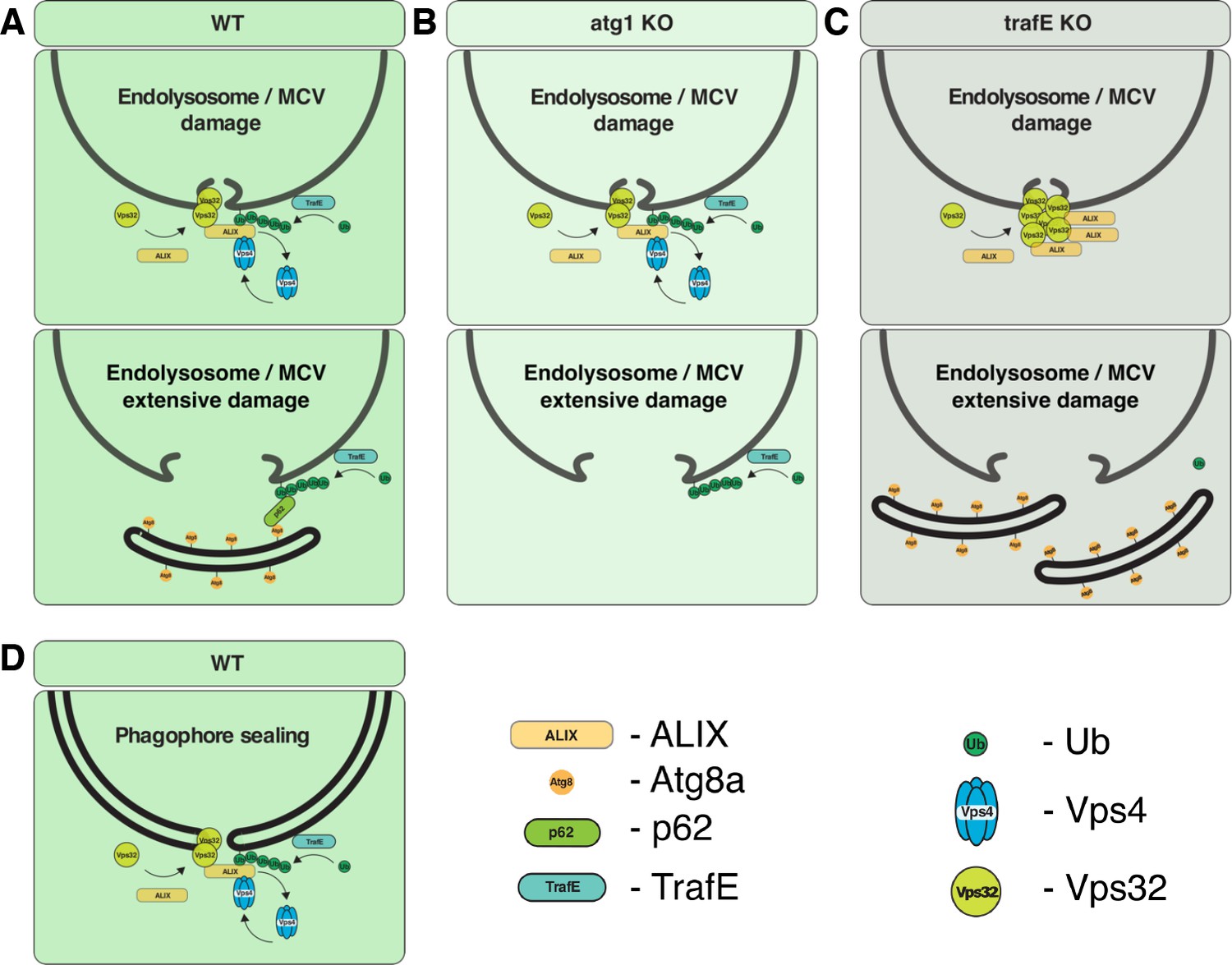
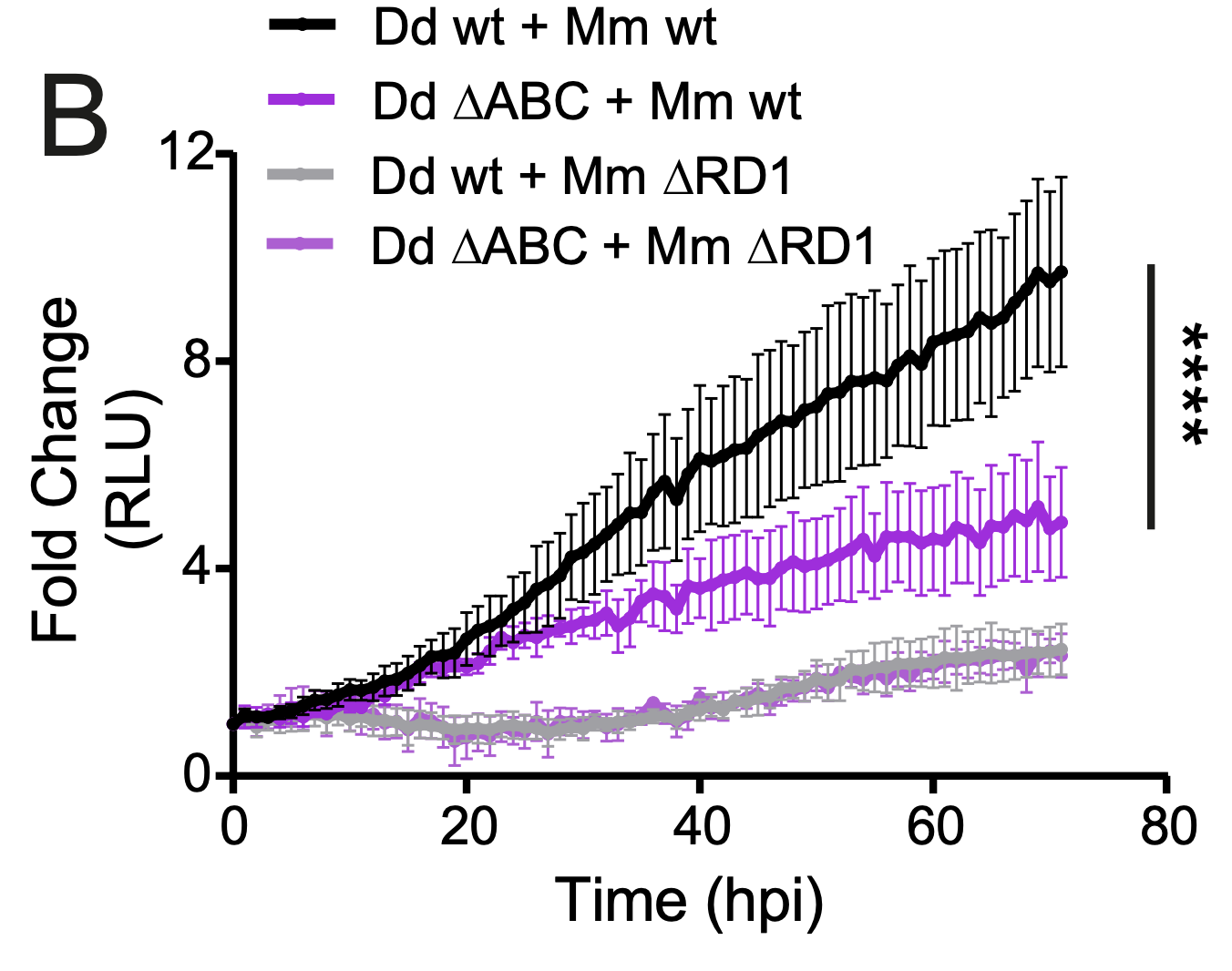
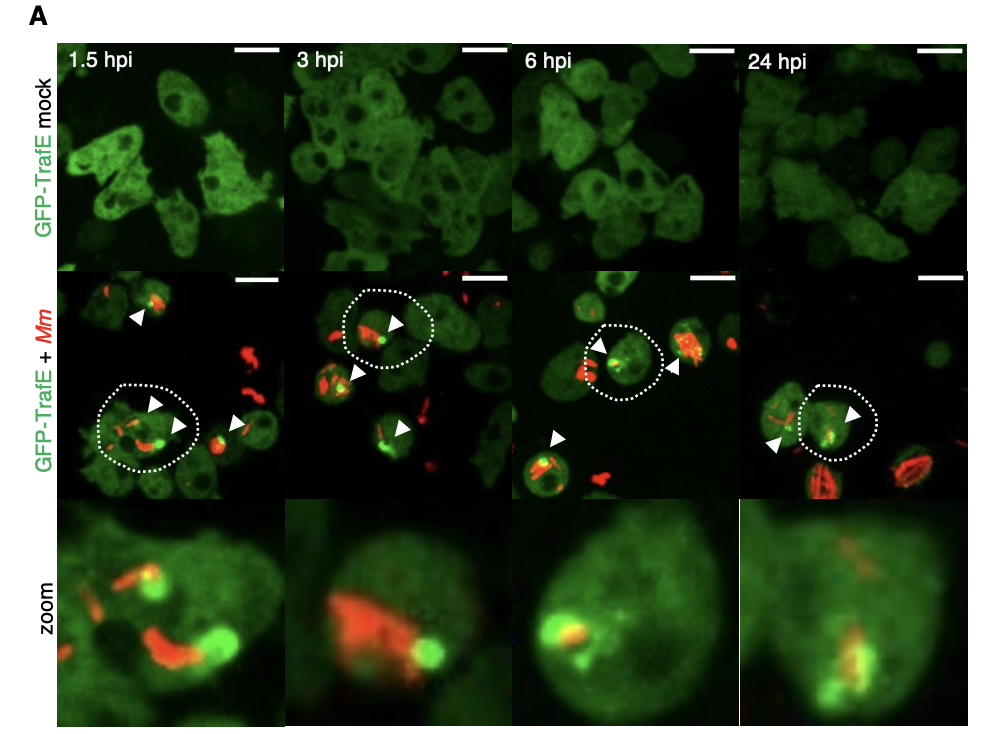
New paper out in eLife: A TRAF-like E3 ubiquitin ligase TrafE coordinates ESCRT and autophagy in endolysosomal damage response and cell-autonomous immunity to Mycobacterium marinum
Cells are perpetually challenged by pathogens, protein aggregates or chemicals, that induce plasma membrane or endolysosomal compartments damage. This severe stress is recognised and controlled by the endosomal sorting complex required for transport (ESCRT) and the autophagy machineries, which are recruited to damaged membranes to either repair or to remove membrane remnants. Yet, insight is limited about how damage is sensed and which effectors lead to extensive tagging of the damaged organelles with signals, such as K63-polyubiquitin, required for the recruitment of membrane repair or removal machineries. To explore the key factors responsible for detection and marking of damaged compartments, we use the professional phagocyte Dictyostelium discoideum. We found an evolutionary conserved E3-ligase, TrafE, that is robustly recruited to intracellular compartments disrupted after infection with Mycobacterium marinum or after sterile damage caused by chemical compounds. TrafE acts at the intersection of ESCRT and autophagy pathways and plays a key role in functional recruitment of the ESCRT subunits ALIX, Vps32 and Vps4 to damage sites. Importantly, we show that the absence of TrafE severely compromises the xenophagy restriction of mycobacteria as well as ESCRT-mediated and autophagy-mediated endolysosomal membrane damage repair, resulting in early cell death.
For the complete story, click here!
MARCH 2023
We have a new postdoctoral fellow in the lab!
Justine Toinon join us for a project on the exploration of the signaling cascades and transcriptional reprogramming in the course of Mycobacterium marinum infection both in Dictyostelium discoideum and in murine microglial cells. Let's give her a warm welcome!
We have a new postdoctoral fellow in the lab!
After his stay last year, Lucas Ceseti decided to come back and join us as a postdoctoral fellow for a project on elucidating the strategies used by X. citri to resist predation by D. discoideum. Let's give him a warm welcome!
November 2022
New collaborative paper: 5-ethyl-2’-deoxyuridine fragilizes Klebsiella pneumoniae outer wall and facilitates intracellular killing by phagocytic cells
Our team collaborated with the team of Prof. Pierre Cosson on this work that allowed the identification of a new antibacterial compound targeting K. pneumoniae and facilitating its killing by Dicty. You can learn a bit more about this story here, or directly read the paper published in PLOS One here.
Klebsiella pneumoniae is the causative agent of a variety of severe infections. Many K. pneumoniae strains are resistant to multiple antibiotics, and this situation creates a need for new antibacterial molecules. K. pneumoniae pathogenicity relies largely on its ability to escape phagocytosis and intracellular killing by phagocytic cells. Interfering with these escape mechanisms may allow to decrease bacterial virulence and to combat infections. In this study, we used Dictyostelium discoideum as a model phagocyte to screen a collection of 1,099 chemical compounds. Phg1A KO D. discoideum cells cannot feed upon K. pneumoniae bacteria, unless bacteria bear mutations decreasing their virulence. We identified 3 non-antibiotic compounds that restored growth of phg1A KO cells on K. pneumoniae, and we characterized the mode of action of one of them, 5-ethyl-2’-deoxyuridine (K2). K2-treated bacteria were more rapidly killed in D. discoideum phagosomes than non-treated bacteria. They were more sensitive to polymyxin and their outer membrane was more accessible to a hydrophobic fluorescent probe. These results suggest that K2 acts by rendering the membrane of K. pneumoniae accessible to antibacterial effectors. K2 was effective on three different K. pneumoniae strains, and acted at concentrations as low as 3 μM. K2 has previously been used to treat viral infections but its precise molecular mechanism of action in K. pneumoniae remains to be determined.
September 2022
A new collaborative work with Dr. Cristina Alvarez-Martinez from the Universidade Estadual de Campinas (Brazil) supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and the Swiss National Science Foundation (SNSF)
Thanks to a research grant, the FAPESP and the SNSF will support a new collaborative work with Dr. Cristina Alvarez-Martinez. This project will aim at deciphering the molecular mechanisms of the Xanthomonas citri resistance to Dictyostelium discoideum predation and the role of the Type VI Secretion System (T6SS) (A) of X. citri in this resistance. Overall, this project will expand our understanding of the role of anti-eukaryotic T6SS and possibly reveal new effector functions and anti-host strategies. This will help to understand better the mechanisms used by X. citri to survive and disseminate in the environment (B).
April 2022
We have a new postdoctoral fellow in the lab!

Sandra Guallar-Garrido join us for a project on the machineries involved in damage sensing and repairing in Dictyostelium discoideum. Let's give her a warm welcome!
January 2022
A new comer in the team for a few months!

Lucas Ceseti, a phD student from the group of Cristina Martinez (https://www.ib.unicamp.br/node/99), joined us for three months in order to work on the relationship between our favorite beast, Dictyostelium discoideum, and the phytopathogen Xanthomonas citri.
"As part of my PhD research performed at Unicamp (Brazil), I went to the Soldati Lab for a 3-month internship to evaluate the phenotypes of Dicty expressing putative T6SS effectors that Xanthomonas citri (Xac) uses to resist against the amoeba. Also during this period, I worked together with the Senior postdoc Céline Michard to establish a potential tool for monitoring translocation from bacteria to Dicty, and much more aiming to understand what happens when Xac meets Dicty." - Lucas Ceseti
November 2021
New preprint from the lab: Disruption of vacuolin microdomains in the host Dictyostelium discoideum increases resistance to Mycobacterium marinum-induced membrane damage and infection
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, manipulates the host phagosome maturation pathway to replicate intracellularly. Mycobacterium marinum, a closely-related species, and Dictyostelium discoideum, a social amoeba and alternative phagocytic host, have been used as models to study host-pathogen interactions occurring during mycobacterial infections. Vacuolins, functional homologues of the mammalian flotillins, organize membrane microdomains and play a role in vesicular trafficking. Various pathogens have been reported to manipulate their membrane association and function. During infection of D. discoideum with M. marinum, Vacuolin C was specifically and highly induced and all three vacuolin isoforms were enriched at the mycobacteria-containing-vacuole (MCV). In addition, absence of vacuolins reduced escape from the MCV and conferred resistance to M. marinum infection. Moreover, ESAT-6, the membrane-disrupting virulence factor of M. marinum, was less associated with membranes when vacuolins were absent. Together, these results suggest that vacuolins are important host factors that are manipulated by mycobacteria to inflict membrane damage and escape from their compartment.
September 2021
Congratulations to our new PhD Lyudmil Raykov!
On September 6th, Lyudmil Raykov successfully defended his phD, entilted "Identification and Characterization of Dictyostelium discoideum Conserved Response Factors Involved in Pathogen Detection and Stress Signal Transduction". A big thank you to the jury members: Prof. Aurélien Roux, Prof. Pierre Cosson and Prof. Félix Randow
July 2021
Our new Methods chapter is out! If you want to learn how to study infection at different scale, you have to read it!
The Dictyostelium discoideum–Mycobacterium marinum host–pathogen system is a well-established and powerful alternative model system to study mycobacterial infections. In this chapter, we will describe three microscopy methods that allow the precise identification and quantification of very diverse phenotypes arising during infection of D. discoideum with M. marinum. First, at the lowest end of the scale, we use the InfectChip, a microfluidic device that enables the long-term monitoring of the integrated history of the infection course at the single-cell level. We use single-cell analysis to precisely map and quantitate the various fates of the host and the pathogen during infection. Second, a high-content microscopy setup was established to study the infection dynamics with high-throughput imaging of a large number of cells at the different critical stages of infection. The large datasets are then fed into a deep image analysis pipeline allowing the development of complex phenotypic analyses. Finally, as part of its life cycle, single D. discoideum amoebae aggregate by chemotaxis to form multicellular structures, which represent ordered assemblies of hundreds of thousands of cells. This transition represents a challenge for the monitoring of infection at multiple scales, from single cells to a true multicellular organism. In order to visualize and quantitate the fates of host cells and bacteria during the developmental cycle in a controlled manner, we can adjust the proportion of infected cells using live FAC-sorting. Then, cells are plated in defined humidity conditions on optical glass plates in order to image large fields, using tile scans, with the help of a spinning disc confocal microscope.
June 2021
We are very happy to share our bioRxiv preprint: The Dictyostelium discoideum E3 ubiquitin ligase TrafE coordinates endolysosomal damage response and cell-autonomous immunity to Mycobacterium marinum.
Cells are perpetually challenged by pathogens, protein aggregates or chemicals, that induce plasma membrane or endolysosomal compartments damage. Endolysosomal perforations are recognised as severe stress, however the mechanisms of the cellular response that ensure quality control, repair and endolysosomal homeostasis are just beginning to be unravelled. The endosomal sorting complex required for transport (ESCRT) and the autophagy machinery are recruited to damaged membranes to either repair or to remove membrane remnants. Crucial element of the endolysosomal damage response (ELDR) are factors that sense damage, paralleled by extensive tagging of the damaged organelles with signals, such as ubiquitin, required for the recruitment of ELDR components. Unattended membrane damage leads to leakage of harmful components including protons and reactive oxygen species that cause cell death. To explore ELDR key factors responsible for detection and marking of damaged compartments we use the professional phagocyte Dictyostelium discoideum. We found an evolutionary conserved E3-ligase TrafE that is robustly recruited to intracellular compartments disrupted after infection with Mycobacterium marinum or after sterile damage caused by chemical components. Importantly, we show that the absence of TrafE severely compromises the xenophagy restriction of bacteria as well as autophagy-mediated and ESCRT-mediated ELDR, resulting in early cell death.
FEBRUARY 2021
Our latest paper is out! Have a look at the effect of Zinc during infection!
Macrophages use diverse strategies to restrict intracellular pathogens, including either depriving the bacteria of (micro)nutrients such as transition metals or intoxicating them via metal accumulation. Little is known about the chemical warfare between Mycobacterium marinum, a close relative of Mycobacterium tuberculosis (Mtb), and its hosts. We use the professional phagocyte Dictyostelium discoideum to investigate the role of Zn2+ during M. marinum infection. We show that M. marinum senses toxic levels of Zn2+ and responds by upregulating one of its isoforms of the Zn2+ efflux transporter CtpC. Deletion of ctpC (MMAR_1271) leads to growth inhibition in broth supplemented with Zn2+ as well as reduced intracellular growth. Both phenotypes were fully rescued by constitutive ectopic expression of the Mtb CtpC orthologue demonstrating that MMAR_1271 is the functional CtpC Zn2+ efflux transporter in M. marinum. Infection leads to the accumulation of Zn2+ inside the Mycobacterium-containing vacuole (MCV), achieved by the induction and recruitment of the D. discoideum Zn2+ efflux pumps ZntA and ZntB. In cells lacking ZntA, there is further attenuation of M. marinum growth, presumably due to a compensatory efflux of Zn2+ into the MCV, carried out by ZntB, the main Zn2+ transporter in endosomes and phagosomes. Counterintuitively, bacterial growth is also impaired in zntB KO cells, in which MCVs appear to accumulate less Zn2+ than in wild-type cells, suggesting restriction by other Zn2+-mediated mechanisms. Absence of CtpC further epistatically attenuates the intracellular proliferation of M. marinum in zntA and zntB KO cells, confirming that mycobacteria face noxious levels of Zn2+.

January 2021
 We have a new postdoctoral fellow in the lab!
We have a new postdoctoral fellow in the lab!
Mélanie Foulon joined us in January for a project on the host-derived lipids: transport and utilization by mycobacteria during their intracellular life! Let's give her a warm welcome!
OCTOBER 2020
Our latest collaboration is out! Dr Mohammad Parhizkar, in the team of Prof Giovanna Di Marzo Serugendo at UNIGE is working on Dictyostelium discoideum as an Inspiration for Higher-Order Emergence in Collective Adaptive Systems and Swarm Robotics.
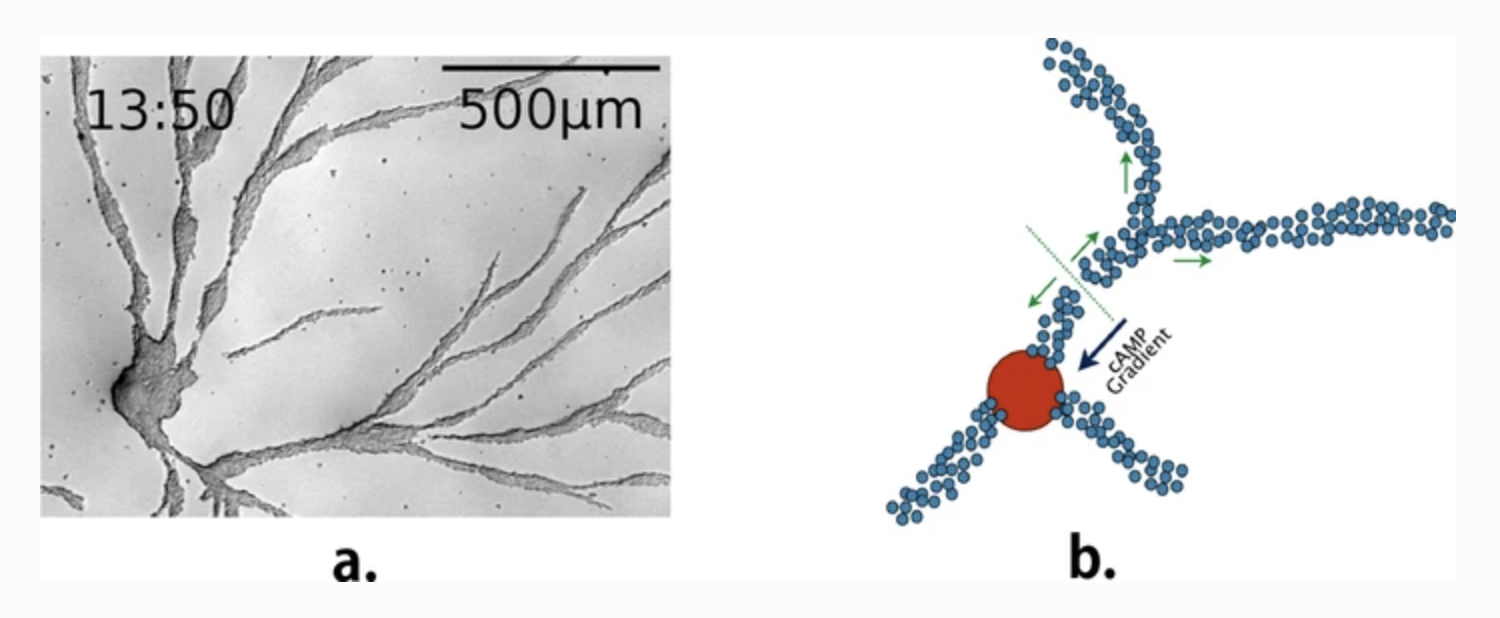
Collective behaviour in nature provides a source of inspiration to engineer artificial collective adaptive systems, due to their mechanisms favouring adaptation to environmental changes and enabling complex emergent behaviour to arise from a relatively simple behaviour of individual entities. As part of our ongoing research, we study the social amoeba Dictyostelium discoideum to derive agent-based models and mechanisms that we can then exploit in artificial systems, in particular in swarm robotics. In this paper, we present a selection of agent-based models of the aggregation phase of D. discoideum, their corresponding biological illustrations and how we used them as an inspiration for transposing this behaviour into swarms of Kilobots. We focus on the stream-breaking phenomenon occurring during the aggregation phase of the life cycle of D. discoideum. Results show that the breakup of aggregation streams depends on cell density, motility, motive force and the concentration of cAMP and CF. The breakup also comes with the appearance of late centres. Our computational results show similar behaviour to our biological experiments, using Ax2(ka) strain. For swarm robotics experiments, we focus on signalling and aggregation towards a centre.
june 2020
We are happy to announce that our collaboration with the lab of Jason King at the University of Sheffield is now out !
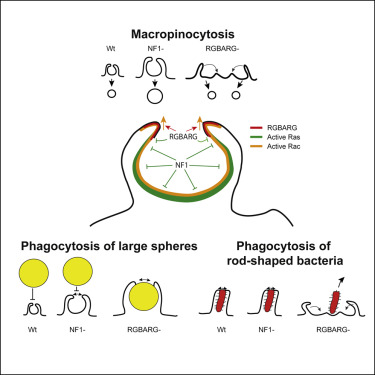
Engulfment of extracellular material by phagocytosis or macropinocytosis depends on the ability of cells to generate specialized cup-shaped protrusions. To effectively capture and internalize their targets, these cups are organized into a ring or ruffle of actin-driven protrusion encircling a non-protrusive interior domain. These functional domains depend on the combined activities of multiple Ras and Rho family small GTPases, but how their activities are integrated and differentially regulated over space and time is unknown. Here, we show that the amoeba Dictyostelium discoideum coordinates Ras and Rac activity using the multidomain protein RGBARG (RCC1, RhoGEF, BAR, and RasGAP-containing protein). We find RGBARG uses a tripartite mechanism of Ras, Rac, and phospholipid interactions to localize at the protruding edge and interface with the interior of both macropinocytic and phagocytic cups. There, we propose RGBARG shapes the protrusion by expanding Rac activation at the rim while suppressing expansion of the active Ras interior domain. Consequently, cells lacking RGBARG form enlarged, flat interior domains unable to generate large macropinosomes. During phagocytosis, we find that disruption of RGBARG causes a geometry-specific defect in engulfing rod-shaped bacteria and ellipsoidal beads. This demonstrates the importance of coordinating small GTPase activities during engulfment of more complex shapes and thus the full physiological range of microbes, and how this is achieved in a model professional phagocyte.
Read our new article on the role of Vacuolins in phagocytic uptake and phagosomal membrane recycling in Dictyostelium discoideum
Flotillins are lipid rafts residents involved in membrane trafficking and recycling of plasma membrane proteins. Dictyostelium discoideum uses phagocytosis to kill, digest and feed on bacteria. It possesses three flotillin-like vacuolins that are strongly associated with membranes and gradually accumulate on maturing phagosomes. Absence of vacuolins reduced adhesion and particle recognition resulting in a drastic reduction in the uptake of various types of particles. This was caused by a block in the recycling of plasma membrane components and the absence of their specific cortex-associated proteins. In addition, absence of vacuolins also impaired phagolysosome biogenesis, without significantly impacting killing and digestion of a range of bacteria. Strikingly, both absence and overexpression of vacuolins induced a strong down-regulation of myosin VII expression, as well as its partner talin A. Episomal expression of myosin VII fully rescued defects in uptake and adhesion, but not in phagosome maturation. These results suggest a dual role for vacuolins: a novel mechanism involving membrane microdomains and myosin VII/talin A in clustering phagosomal receptors and adhesion molecules at the plasma membrane, and a role in phagolysosomal biogenesis.
MAY 2020
We are happy to announce that our collaboration with the lab of Falk Hillmann at the HKI in Jena, and especially with Iuliia Viediernikova who came two month in 2017 with an EMBO fellowship was fruitful and the article is now out !
The human-pathogenic fungus Aspergillus fumigatusis a ubiquitous saprophyte that causes fatal lung infections in immunocompromised individuals. Following inhalation, conidia are ingested by innate immune cells and can arrestphagolysosome maturation. How this virulence trait could have been selected for innatural environments is unknown. Here, we found that surface exposure of thegreen pigment 1,8-dihydroxynaphthalene-(DHN)-melanin can protect conidia from phagocytic uptake and intracellular killing by the fungivorous amoeba Protostelium aurantium and delays its exocytosis from the non fungivorous species Dictyostelium discoideum. To elucidate the antiphagocytic properties of the surface pigment, we followed the antagonistic interactions of A. fumigatus conidia with the amoebae in real time. For both amoebae, conidia covered with DHN-melanin were internalizedat far lower rates than were seen with conidia lacking the pigment, despite high rates of initial attachment to non killing D. discoideum. When ingested by D. discoideum, the formation of nascent phagosomes was followed by transient acidification of phagolysosomes, their subsequent neutralization, and, finally, exocytosis of the conidia. While the cycle was completed in less than 1 h for unpigmented conidia, the process was significantly prolonged for conidia covered with DHN-melanin, leading to an extended intracellular residence time. At later stages of this cellular infection, pigmented conidia induced enhanced damage to phagolysosomes and infected amoebae failed to recruit the ESCRT (endosomal sorting complex required for transport) membrane repair machinery or the canonical autophagy pathway to defend against the pathogen, thus promoting prolonged intracellular persistence in the host cell and the establishment of a germination niche in this environmental phagocyte.
March 2020
New Post-doctoral fellow in the lab!
Céline Michard joined the Soldati Lab in March. Please give her a warm welcome! She will work on the mechanistic aspect of host membrane repair during mycobacterial infection.
FEBRUARY 2020
We have a new MSc in the lab!
Krikor Bared Eblighatian is joining us. He will work on Flipper probes to label the MCV in D. discoideum and to measure membrane tension upon hyperosmotic shock, upon sterile damage and during M. marinum infection
Welcome to our new PhD student!
Davide D'Amico joined the team in Februrary. He will work on the host response to vacuolar escape and better dissect the role of discoidins in the autophagic pathway. Let's give him a warm welcome!
Finally, the core findings of our collaborative study supported by SNF Sinergia and SystemsX.ch HostPathX grants are published. Enjoy !!
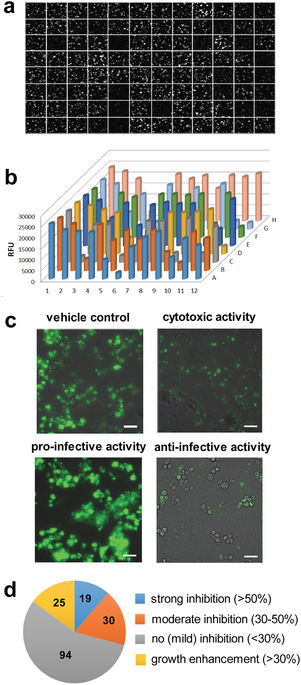
Tubercular Mycobacteria and Legionella pneumophila are the causative agents of potentially fatal respiratory diseases due to their intrinsic pathogenesis but also due to the emergence of antibiotic resistance that limits treatment options. The aim of our study was to explore the antimicrobial activity of a small ligand-based chemical library of 1255 structurally diverse compounds. These compounds were screened in a combination of three assays, two monitoring the intracellular growth of the pathogenic bacteria, Mycobacterium marinum and L. pneumophila, and one assessing virulence of M. marinum. We set up these assays using two amoeba strains, the genetically tractable social amoeba Dictyostelium discoideum and the free-living amoeba Acanthamoeba castellanii. In summary, 64 (5.1%) compounds showed anti-infective/anti-virulence activity in at least one of the three assays. The intracellular assays hit rate varied between 1.7% (n = 22) for M. marinum and 2.8% (n = 35) for L. pneumophila with seven compounds in common for both pathogens. In parallel, 1.2% (n = 15) of the tested compounds were able to restore D. discoideum growth in the presence of M. marinum spiked in a lawn of food bacteria. We also validated the generality of the hits identified in the A. castellanii–M. marinum anti-infective screen using the D. discoideum–M. marinum host–pathogen model. The characterization of anti-infective and antibacterial hits in the latter infection model revealed compounds able to reduce intracellular growth more than 50% at 30 μM. Moreover, the chemical space and physico-chemical properties of the anti-M. marinum hits were compared to standard and candidate Mycobacterium tuberculosis (Mtb) drugs using ChemGPS-NP. A principle component analysis identified separate clusters for anti-M. marinum and anti-L. pneumophila hits unveiling the potentially new physico-chemical properties of these hits compared to standard and candidate M. tuberculosis drugs. Our studies underscore the relevance of using a combination of low-cost and low-complexity assays with full 3R compliance in concert with a rationalized focused library of compounds to identify new chemical scaffolds and to dissect some of their properties prior to taking further steps toward compound development.
 New MSc student in the lab!
New MSc student in the lab!
Angélique Perret starts today her MSc with us. Let's give her a warm welcome! She will compare the infection process of M. marinum in D. discoideum and in BV-2 murine microglial cells!
January 2020
Read our new collaborative paper with the Hibli Lab in Zürich!
Mycobacterium marinum is a model organism for pathogenic Mycobacterium species, including Mycobacterium tuberculosis, the causative agent of tuberculosis. These pathogens enter phagocytes and replicate within the Mycobacterium‐containing vacuole (MCV), possibly followed by vacuole exit and growth in the host cell cytosol. Mycobacteria release siderophores called mycobactins to scavenge iron, an essential, yet poorly soluble and available micro‐nutrient. To investigate the role of M. marinum mycobactins, we purified by organic solvent extraction and identified by mass spectrometry the lipid‐bound mycobactin (MBT) and the water‐soluble variant carboxymycobactin (cMBT). Moreover, we generated by specialized phage transduction a defined M. marinum ΔmbtB deletion mutant predicted to be defective for mycobactin production. The M. marinum ΔmbtB mutant strain showed a severe growth defect in broth and phagocytes, which was partially complemented by supplying the mbtB gene on a plasmid. Furthermore, purified Fe‐MBT or Fe‐cMBT improved the growth of wild‐type as well as ΔmbtB mutant bacteria on minimal plates, but only Fe‐cMBT promoted the growth of wild‐type M. marinum during phagocyte infection. Finally, the intracellular growth of M. marinum ΔmbtB in Acanthamoeba castellanii amoebae was restored by co‐infection with wild‐type bacteria. Our study identifies and characterizes the M. marinum MBT and cMBT siderophores and reveals the requirement of mycobactins for extra‐ and intracellular growth of the pathogen.
This paper can be read here: Knobloch P, Koliwer-Brandl H, Arnold FM, Hanna N, Gonda I, Adenau S, Personnic N, Barisch C, Seeger MA, Soldati T, Hilbi H (2020). Mycobacterium marinum produces distinct mycobactin and carboxymycobactin siderophores to promote growth in broth and phagocytes.
Cell Microbiol. doi: 10.1111/cmi.13163
Join us on January 16 & 17, 2020 at the School of Physics in Geneva!
Come to learn more about the science going on in the Chemistry and Biochemistry department! We have 5 outstanding international speaker plus 14 junior speakers! Among them, our own PhD Student, Lyudmil Raykov, will talk to you about "Conserved microbial restriction factors in Dictyostelium discoideum cell-autonomous immunity"!
Read our first collaborative paper of a hopefully long series for 2020!
Mycobacterium bovis is the causative agent of bovine tuberculosis and the predominant cause of zoonotic tuberculosis in people. Bovine tuberculosis occurs in farmed cattle but also in a variety of wild animals, which form a reservoir of infection. Although direct transmission of tuberculosis occurs between mammals, the low frequency of contact between different host species and abundant shedding of bacilli by infected animals suggests an infectious route via environmental contamination. Other intracellular pathogens that transmit via the environment deploy strategies to survive or exploit predation by environmental amoebae. To explore if M. bovis has this capability, we investigated its interactions with the soil and dung-dwelling amoeba, Dictyostelium discoideum. We demonstrated that M. bovis evades phagocytosis and destruction by D. discoideum and actively transits through the amoeba using the ESX-1 Type VII Secretion System as part of a programme of mechanisms, many of which have been co-opted as virulence factors in the mammalian host. This capacity of M. bovis to utilise an environmental stage between mammalian hosts may enhance its transmissibility. In addition, our data provide molecular evidence to support an evolutionary role for amoebae as training grounds for the pathogenic M. tuberculosis complex.
 This paper can be read here: Butler RE, Smith AA, Mendum TA, Chandran A, Wu H, Lefrançois L, Chambers M, Soldati T, Stewart GR. (2020) Mycobacterium bovis uses the ESX-1 Type VII secretion system to escape predation by the soil-dwelling amoeba Dictyostelium discoideum.
This paper can be read here: Butler RE, Smith AA, Mendum TA, Chandran A, Wu H, Lefrançois L, Chambers M, Soldati T, Stewart GR. (2020) Mycobacterium bovis uses the ESX-1 Type VII secretion system to escape predation by the soil-dwelling amoeba Dictyostelium discoideum.
The ISME Journal. doi: 10.1038/s41396-019-0572-z.
NOVember 2019
A SNSF Sinergia grant has been assigned to our collaborative project "An in silico and chemo-biological approach to identify anti-infective and pro-metabolic natural products"

Image ©UNIGE
"The recent rapid innovations made in chemical profiling may lead to a change of paradigm in natural products (NP) research. Especially the increasing amount of accurate metabolome data that can be acquired on massive sample sets notably through advanced liquid chromatography mass spectrometry (LC/MS) provides a large amount of information on plant composition at an unprecedented level of precision. Being able to exhaustively identify the structures of all possible metabolites within any given natural extract in parallel to advanced bioassays opens the possibility to undertake NP-based drug discovery studies and guide the targeted isolation direct NP synthesis of lead compounds using a more comprehensive and effective strategy. In this project we will leverage these possibilities to develop an innovative integrated in silico and chemo-biological approach to efficiently identify anti-infective and pro-metabolic NPs. This will serve to address two important pathologies with highly unmet clinical needs namely tuberculosis and obesity as well as their associated co-morbidities. We have selected these two different pathologies as they share several common features that can be leveraged to identify bioactive extracts and compound classes which can be used to target directly or indirectly these diseases. This is based on the fact that lipid droplet formation morphology and functionality as well as signalling via reactive oxygen species are important both for host response to Mycobacterium tuberculosis infection as well as for systemic metabolic control through adipose tissue functionality. The combination of bioassays aiming at monitoring the anti-infective and pro-metabolic activities of natural extracts and compounds is expected to yield comprehensive information on the activity of different molecules under specific paradigms. Based on these combined studies downstream refined assays for specific cellular functionality coupled to mechanistic studies to delineate the NPs’ mode of action will be conducted. To achieve the goals of this proposal we will utilize a unique library of more than 15500 plant parts that is made available by a partnership with Pierre Fabre. This will give us the possibility to apply the approach on a massive set of samples while benefitting from Pierre Fabre’s botanical expertise to conduct the research in full compliance with international regulations regarding the access to biodiversity. We hypothesise that through massive high-quality MS data combined with the use of adapted computational metrics we will achieve an unprecedented level of precision which will allow us to obtain information on active scaffolds by targeted isolation of NPs of interest and the synthesis of analogues with optimized activity profiles. These identified scaffolds and analogues can subsequently serve to generate new treatment strategies and/or accompany existing treatments for combating both tuberculosis infection as well as obesity and their associated co-morbidities."
This project is a collaboration between our group and the groups of Prof. Jean-Luc Wolfender (UNIGE), Prof. Erik M. Carreira (ETH Zurich) and Prof. Christian Wolfrum (ETH Zurich).
We will be present at the "Cell Dynamics: Host-Pathogen Interface" meeting next 17-20 May 2020 in Surrey, UK. Will you join us?
Find all info and registration process here.
October 2019
FEBS3+ LS2 Annual Meeting, 13-14 February 2020 in Zurich!
As every year, we will be present at the LS2 Annual Meeting. For the first time in 2020, the LS2 Annual Meeting welcomes the German Society for Biochemistry and Molecular Biology (GBM) & the Austrian Association of Molecular Life Sciences and Biotechnology (ÖGMBT) as co-organizing societies under the FEBS3+ umbrella.
Among many interesting scientific & career sessions, there will be a dedicated symposium to the Cell Biology of Infection! Will you join us next February in Zurich? You can find all the information about the meeting and how to register here: https://annual-meeting.ls2.ch.

Honoured to receive the 2019 3R Award of the University of Geneva!
The prize is awarded in recognition of our project entitled "Antimycobacterial drug discovery using Mycobacteria-infected amoebae identifies anti-infectives and new molecular targets”. Thank you to the University and to my group, who help me contributing to the 3Rs principle!
The press release about our prize can be read here.
Additional reading and videos related to the prize:
- Jump-To-Science (UNIGE)
- TV Léman Bleu
- Techno-Science.net
- Bote
- Mirage News
- Blick
- Nau.ch
- Schweizer Bauer
- Volksblatt
- Tierwelt
- bz
- derStandard
- swissinfo
- Limmattaler Zeitung
- Kleine Zeitung
Registration is open for "Technologie génétique! Et vous? Discutons-en!"
May the recent innovations in genetic engineering impact our everyday life? What are the opportunities, limits, risks, and ethical considerations and decisions that concern us as a society? Participate in an open discussion on this topical issue beyond decades of controversy, discover other points of view and make your own opinion. Organized by the Forum for Genetic Research of the Swiss Academy of Sciences (SCNAT) with the support of the Federal Office for the Environment.
When: November 20, 2019, 14:45 - 19:30.
Where: Impact Hub, Lausanne (CH)
More info and online registration here: http://ow.ly/nDYb50wGnwJ.

Image: Forum Genforschung
September 2019
Welcome to our visitor Sandra Guallar-Garrido! 
Sandra is a PhD student from Universitat Autònoma de Barcelona whose main project is based on studying the influence of culture medium in the antitumoral effect of mycobacteria in bladder cancer. She will be in our lab for 3 months thanks to an EMBO Short-Term Fellowship to study the impact of the joint effect of PDIM and ESAT-6 of Mycobacterium marinum during Dictyostelium discoideum infection.
july 2019
We are hiring!
The group is ready to welcome excellent, very motivated PhD candidates and outstanding postdoctoral fellows with interest and expertise in various fields of cell biology, biochemistry and genetics, with an emphasis on cellular microbiology, bacteriology and host-pathogen interactions. For more information on how to apply, please click here.
APRIL 2019
Our collaboration with the Wolfender group is out!
The water decoction of Combretum aculeatum aerial parts is traditionally used in Senegal to treat tuberculosis. The targeted isolation of its antimycobacterial compounds, carried out in this work, supports its use.
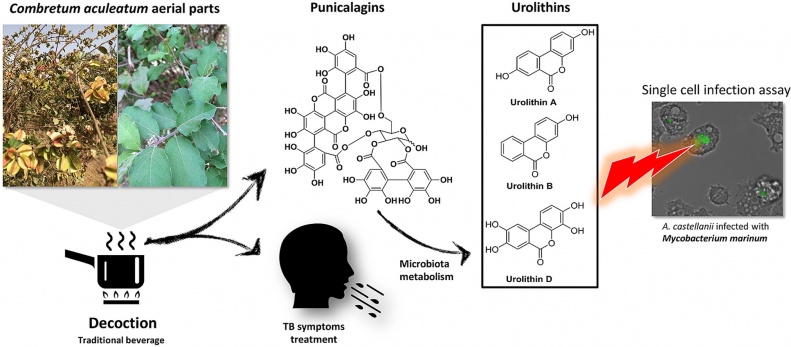
This paper can be read here: Diop EA, Queiroz EF, Marcourt L, Kicka S, Rudaz S, Diop T, Soldati T, Wolfender JL. Antimycobacterial activity in a single-cell infection assay of ellagitannins from Combretum aculeatum and their bioavailable metabolites. J Ethnopharmacol 2019, doi: 10.1016/j.jep.2019.111832.
Join us on May 3, 2019 at University of Zurich!
As Editor in Chief of the Cellular Microbiology journal, I am co-organising the first Cell Microbiology/UZH joint Symposium, where we will celebrate the Cellular Microbiology's 20th anniversary! 20 years of outstanding discoveries from virus to bacteria & fungi pathogens, and from amoebae to animal & plant hosts. It is free and no registration is needed!
Check the program here: Cellular Microbiology Symposium program.pdf
March 2019
New lab member!
Welcome to our new PhD student Jahn Nitschke. Jahn will characterise novel anti-tuberculosis compounds and will run image-based analysis of collective streaming behaviour in Dictyostelium.

FEBRUARY 2019
Our Editorial on the Frontiers in Cellular and Infection Microbiology research topic "Amoebae as Host Models to Study the Interaction with Pathogens" is out
Thank you to all the 70 authors who have contributed to make this research topic a success!
The articles included in this Research Topic highlight the potential of amoebae as model hosts to study the interaction with a wide range of pathogenic microorganisms. The editorial can be read here: Thewes S, Sodati T, Eichinger L. Editorial: Amoebae as Host Models to Study the Interaction with Pathogens. Front. Cell. Infect. Microbiol. 2019, doi: 10.3389/fcimb.2019.00047
New collaborative article!
We demonstrate that PIKfyve-catalysed phosphoinositide production is essential during early phagosome maturation, protecting host cells from diverse pathogenic microbes. Have a look!
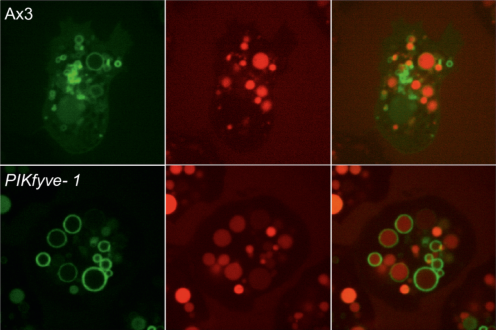
This paper can be read here: Buckley CM, Heath VL, Guého A, Bosmani C, Knobloch P, Sikakana P, Personnic N, Dove SK, Michell RH, Meier R, Hilbi H, Sodati T, Insall RH, King JS. PIKfyve/Fab1 is required for efficient V-ATPase and hydrolase delivery to phagosomes, phagosomal killing, and restriction of Legionella infection. PLoS Pathog 2019, doi: 10.1371/journal.ppat.1007551
JANUARY 2019
Our new paper in collaboration with the group of Hubert Hilbi is now out!
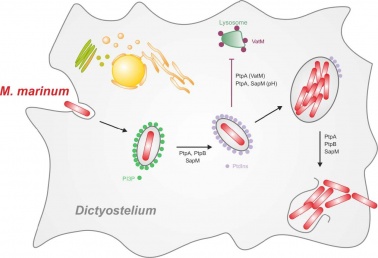
december 2018
We finish 2018 with the publication of Ana's paper! We uncovered how ESCRT and autophagy machineries cooperate to repair the ESX-1-dependent damage at the Mycobacterium-containing vacuole.
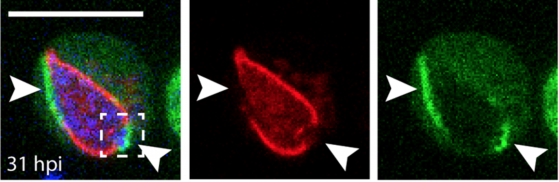
The study has been highlighted in:
Cristina defended her thesis!
Congratulations, Dr. Bosmani! So much data, so well performed and explained. Definitively, a very well deserved PhD.

november 2018
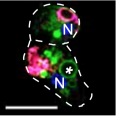 Caroline's paper on the Dictyostelium ZnT zinc transporters has been accepted and published!
Caroline's paper on the Dictyostelium ZnT zinc transporters has been accepted and published!
September 2018

New MSc student in the lab!
Kevin Assoumou starts today his MSc with us. Let's give him a warm welcome!
july 2018
Functions of the Dictyostelium LIMP-2/CD36 homologues in bacteria uptake, phagolysosome biogenesis and host cell defence. Check out our recent paper in Journal of Cell Science:
Phagocytic cells take up, kill and digest microbes by a process called phagocytosis. To this end these cells bind the particle, rearrange their actin cytoskeleton, and orchestrate transport of digestive factors to the particle-containing phagosome. The mammalian lysosomal membrane protein LIMP-2 and CD36, members of the class B of scavenger receptors, play a crucial role in lysosomal enzyme trafficking and uptake of mycobacteria, respectively, and generally in host cell defences against intracellular pathogens. Here, we show that the Dictyostelium discoideum LIMP-2 homologue LmpA regulates phagocytosis and phagolysosome biogenesis. The lmpA knockdown mutant is highly affected in actin-dependent processes such as particle uptake, cellular spreading and motility. Additionally, the cells are severely impaired in phagosomal acidification and proteolysis, likely explaining the higher susceptibility to infection with the pathogenic bacterium Mycobacterium marinum, a close cousin of the human pathogen Mycobacterium tuberculosis. Furthermore, we bring evidence that LmpB is a functional homologue of CD36 and specifically mediates uptake of mycobacteria. Altogether, these data indicate a role for LmpA and LmpB, ancestors of the LIMP-2/CD36 family, in lysosome biogenesis and host cell defence.
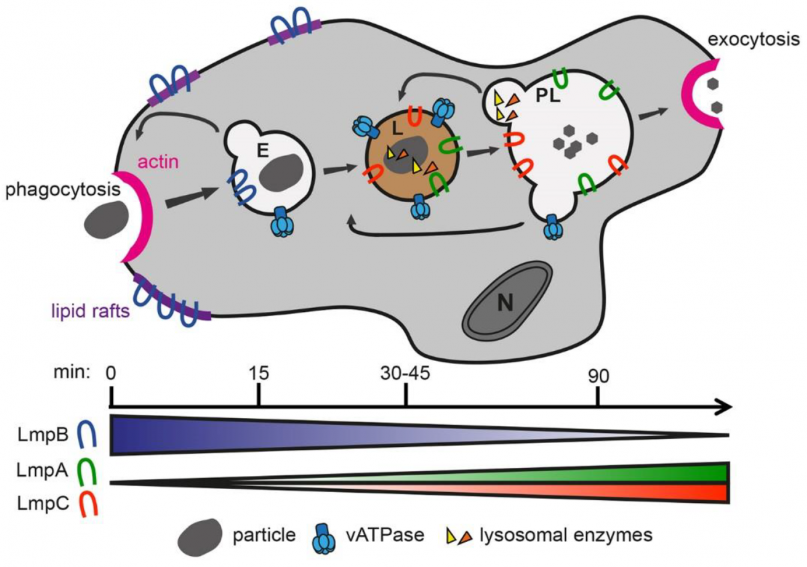
This paper can be read here: , , , , , , , , ,
The study has been highlighted in:
june 2018
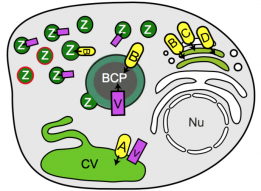
New preprint from the lab:
Professional phagocytes have developed an extensive repertoire of autonomous immunity strategies to ensure killing of bacteria. Besides phagosome acidification and the generation of reactive oxygen species, deprivation of nutrients and the lumenal accumulation of toxic metals are essential to kill ingested bacteria or inhibit growth of intracellular pathogens. We use the soil amoeba Dictyostelium discoideum, a professional phagocyte that digests bacteria for nutritional purposes, to decipher the role of zinc poisoning during phagocytosis of non-pathogenic bacteria and visualize the temporal and spatial dynamics of compartmentalized, free zinc using fluorescent probes. Immediately after particle uptake, zinc is delivered to phagosomes by fusion with 'zincosomes' of endosomal origin, but also by the action of one or more zinc transporters. We localize the four Dictyostelium ZnT transporters to endosomes, the contractile vacuole and the Golgi apparatus, and study the impact of zntknockouts on zinc homeostasis. Finally, we show that zinc is delivered into the lumen of Mycobacterium smegmatis-containing vacuoles, and that Escherichia coli deficient in the zinc efflux P1B-type ATPase ZntA is killed faster than wild type bacteria.
Congratulations Louise Lefrançois, for winning a talk prize at the 2nd NTM European Conference!
MAY 2018
We are very happy to share our bioRxiv preprint "ESCRT and autophagy cooperate to repair ESX-1-dependent damage to the Mycobacterium-containing vacuole".
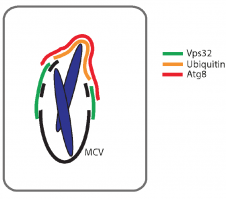
We believe this is a landmark manuscript, hopefully published soon! It can be read here https://doi.org/10.1101/334755.
Thierry Soldati has been elected as EMBO (European Molecular Biology Organisation) Member
Joining a group of more than 1800 of the best researchers in Europe and around the world.
Congratulations, Thierry!

http://www.embo.org/news/press-releases/2018/62-life-scientists-elected-as-embo-members
March 2018
March brings us another publication, this time in collaboration with P. Cosson (University of Geneva) and M. Leippe (University of Kiel). We propose the Apl protein family of Dictyostelium discoideum as antimicrobial effectors.
Due to their archaic life style and microbivor behavior, amoebae may represent a source of antimicrobial peptides and proteins. The amoebic protozoon Dictyostelium discoideum has been a model organism in cell biology for decades and has recently also been used for research on host-pathogen interactions and the evolution of innate immunity. In the genome of D. discoideum, genes can be identified that potentially allow the synthesis of a variety of antimicrobial proteins. However, at the protein level only very few antimicrobial proteins have been characterized that may interact directly with bacteria and help in fighting infection of D. discoideum with potential pathogens. Here, we focus on a large group of gene products that structurally belong to the saposin-like protein (SAPLIP) family and which members we named provisionally Apls (amoebapore-like peptides) according to their similarity to a comprehensively studied antimicrobial and cytotoxic pore-forming protein of the protozoan parasite Entamoeba histolytica. We focused on AplD because it is the only Apl gene that is reported to be primarily transcribed further during the multicellular stages such as the mobile slug stage. Upon knock-out (KO) of the gene, aplD− slugs became highly vulnerable to virulent Klebsiella pneumoniae. AplD− slugs harbored bacterial clumps in their interior and were unable to slough off the pathogen in their slime sheath. Re-expression of AplD in aplD− slugs rescued the susceptibility toward K. pneumoniae. The purified recombinant protein rAplD formed pores in liposomes and was also capable of permeabilizing the membrane of live Bacillus megaterium. We propose that the multifarious Apl family of D. discoideum comprises antimicrobial effector polypeptides that are instrumental to interact with bacteria and their phospholipid membranes. The variety of its members would allow a complementary and synergistic action against a variety of microbes, which the amoeba encounters in its environment.
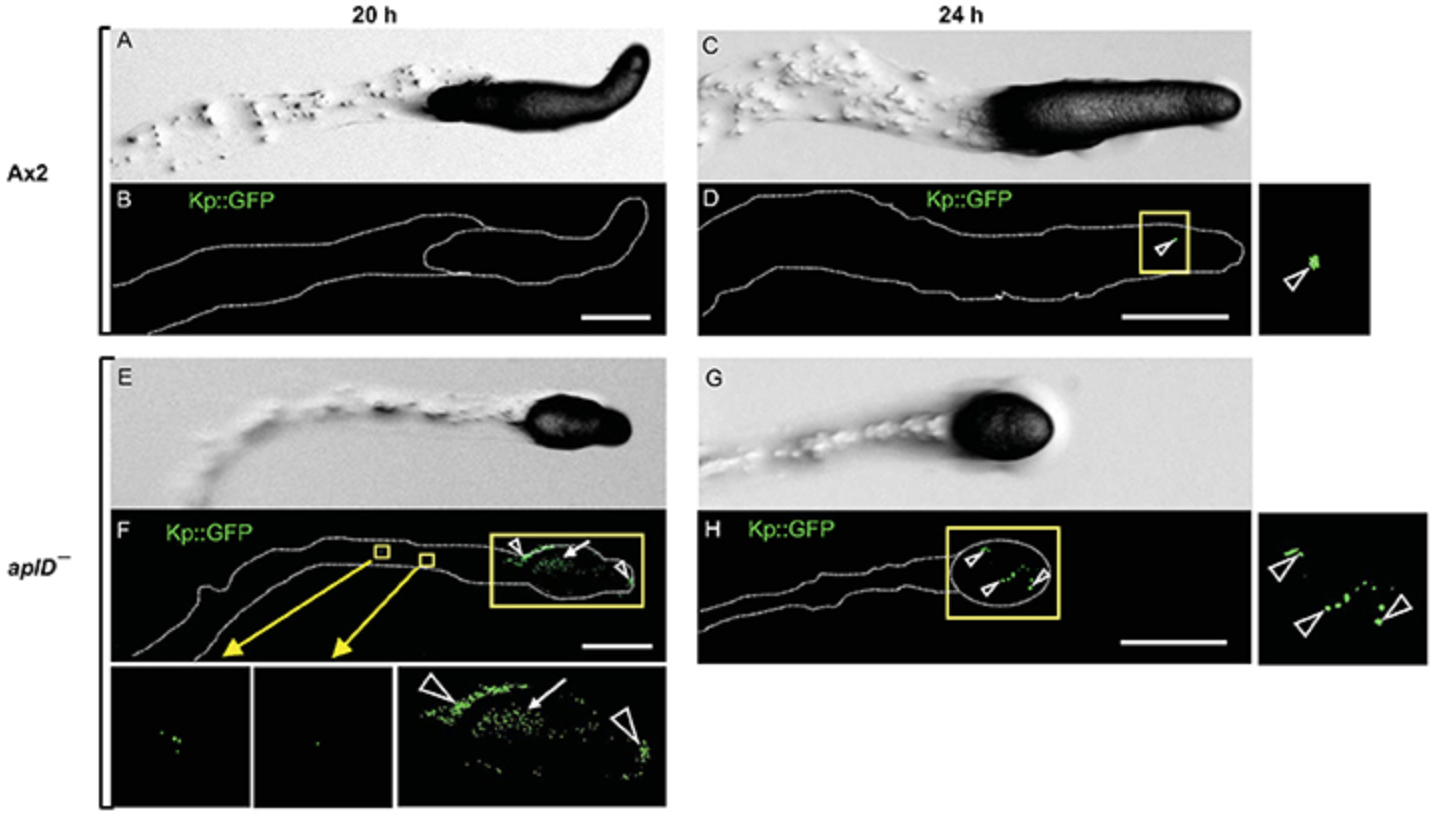
This paper can be read here: Dhakshinamoorthy R, Bitzhenner M, Cosson P, Soldati T, Leippe M. The saponin-like protein AplD displays pore-forming activity and participates in defense against bacterial infection during a multicellular stage of Dictyostelium discoideum. Front Cell Infect Microbiol 2018 | DOI:10.3389/fcimb.2018.00073
Glad to announce that our new paper in collaboration with L. Scapozza (University of Geneva), GS. Besra (University of Birmingham), JAG. Cox (Aston University) and L. Ballell (GSK) is now online in Scientific Reports. We identified anti-infective compounds and new mycobacterial targets for treatment.
Tuberculosis remains a serious threat to human health world-wide, and improved efficiency of medical treatment requires a better understanding of the pathogenesis and the discovery of new drugs. In the present study, we performed a whole-cell based screen in order to complete the characterization of 168 compounds from the GlaxoSmithKline TB-set. We have established and utilized novel previously unexplored host-model systems to characterize the GSK compounds, i.e. the amoeboid organisms D. discoideum and A. castellanii, as well as a microglial phagocytic cell line, BV2. We infected these host cells with Mycobacterium marinum to monitor and characterize the anti-infective activity of the compounds with quantitative fluorescence measurements and high-content microscopy. In summary, 88.1% of the compounds were confirmed as antibiotics against M. marinum, 11.3% and 4.8% displayed strong anti-infective activity in, respectively, the mammalian and protozoan infection models. Additionally, in the two systems, 13–14% of the compounds displayed pro-infective activity. Our studies underline the relevance of using evolutionarily distant pathogen and host models in order to reveal conserved mechanisms of virulence and defence, respectively, which are potential “universal” targets for intervention. Subsequent mechanism of action studies based on generation of over-expresser M. bovis BCG strains, generation of spontaneous resistant mutants and whole genome sequencing revealed four new molecular targets, including FbpA, MurC, MmpL3 and GlpK.
This paper can be read here: Trofimov V, Kicka S, Mucaria S, Hanna N, Ramon-Olayo F, Vela-Gonzalez Del Peral L, Lelièvre J, Ballell L, Scapozza L, Besra GS, Cox JAG, Soldati T. Antimycobacterial drug discovery using Mycobacteria-infected amoebae identifies anti-infectives and new molecular targets. Sci Rep 2018, 8:3939 | DOI:10.1038/s41598-018-22228-6
The study has been highlighted in:
January 2018
We say "until the next time" to Joe Dan, who has been a maître assistant in our laboratory, and a very good friend, for the last 3 years and a half. We will miss your scientific brainstormings during the lab meetings.
The very best for your foreseeable future, Mr. Dunn! 
Our second review of the year was just published! "When Dicty Met Myco, a (Not So) Romantic Story about One Amoeba and Its Intracellular Pathogen".
In this review, we focus on the use of Dictyostelium discoideum as a phagocyte model for the study of mycobacterial infections, in particular Mycobacterium marinum. We look in detail at the intracellular cycle of M. marinum, from its uptake by D. discoideum to its active or passive egress into the extracellular medium. In addition, we introduce the methods and specific tools that have been used so far to monitor the D. discoideum—M. marinum interaction.
Find here our F1000 recommendation of the article "Intracellular Salmonella induces aggrephagy of host endomembranes in persistent infections", published in Autophagy by the laboratory of F García-Del Portillo.

New review by our lab: "Eat Prey, Live: Dictyostelium discoideum As a Model for Cell-Autonomous Defenses".
Here, we discuss the advantages and relevance of Dictyostelium discoideum as a model phagocyte to study cell-autonomous defenses. We cover the antimicrobial functions of phagocytosis and autophagy and describe the processes that create a microbicidal phagosome. We also describe microbial interference with these defenses and highlight observations made first in D. discoideum. Finally, we discuss galectins, TNF receptor-associated factors, tripartite motif-containing proteins, and signal transducers and activators of transcription, microbial restriction factors initially characterized in mammalian phagocytes that have either homologs or functional analogs in D. discoideum.
December 2017
Our new paper "Survey on medicinal plants traditionally used in Senegal for the treatment of tuberculosis (TB) and assessment of their antimycobacterial activity", in collaboration with Prof. Wolfender, is out!
In West Africa, populations are used to taking traditional medicine as a first aid against common health problems. In this aspect, many plants are claimed to be effective in the treatment of Tuberculosis (TB), which according to the World Health Organization (WHO) remains one of the world’s deadliest communicable diseases.The main aim of this study was to identify plants used to treat TB-symptoms by the population of Senegal and to evaluate their possible concomitant use with clinically approved TB-drugs. This approach allowed the selection of plants effectively used in traditional medicine. In order to verify if the usage of some of these plants can be rationalized, the activity of their traditional preparations was assessed with both an intracellular and extracellular antimycobacterial host-pathogen assays.
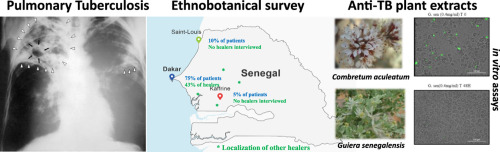
This paper can be read here: Diop EA, Queiroz EF, Kicka S, Rudaz S, Diop T, Soldati T, Wolfender JL. Survey on medicinal plants traditionally used in Senegal for the treatment of tuberculosis (TB) and assessment of their antimycobacterial activity. J Ethnopharmacol 2017, doi: 10.1016/j.jep.2017.12.037.
http://www.sciencedirect.com/science/article/pii/S0378874117322602
We published the biochemical characterization and the first crystal structure of DdPoxA, a secreted heme peroxidase from Dictyostelium discoideum. Find all the information in our new paper in JBC.
Oxidation of halides and thiocyanate by heme peroxidases to antimicrobial oxidants is an important cornerstone in the innate immune system of mammals. Interestingly, phylogenetic and physiological studies suggest that homologous peroxidases are already present in mycetozoan eukaryotes such as Dictyostelium discoideum. This social amoeba kills bacteria via phagocytosis for nutrient acquisition at its single-cell stage and for antibacterial defense at its multicellular stages. Here we demonstrate that peroxidase A from D. discoideum (DdPoxA) is a stable, monomeric, glycosylated and secreted heme peroxidase with homology to mammalian peroxidases. The first crystal structure (2.5 Å resolution) of a mycetozoan peroxidase of this superfamily shows the presence of a posttranslationally-modified heme with one single covalent ester bond between the 1-methyl heme substituent and E236. The metalloprotein follows the halogenation cycle, whereby Compound I oxidizes iodide and thiocyanate at high (> 108 M-1 s-1) and bromide at very low rates. It is demonstrated that DdPoxA is upregulated and likely secreted at late multicellular development stages of D. discoideum when migrating slugs differentiate into fruiting bodies that contain persistent spores on top of a cellular stalk. Expression of DdPoxA is shown to restrict bacterial contamination of fruiting bodies. Structure and function of DdPoxA are compared to evolutionary related mammalian peroxidases in the context of non specific immune defense.
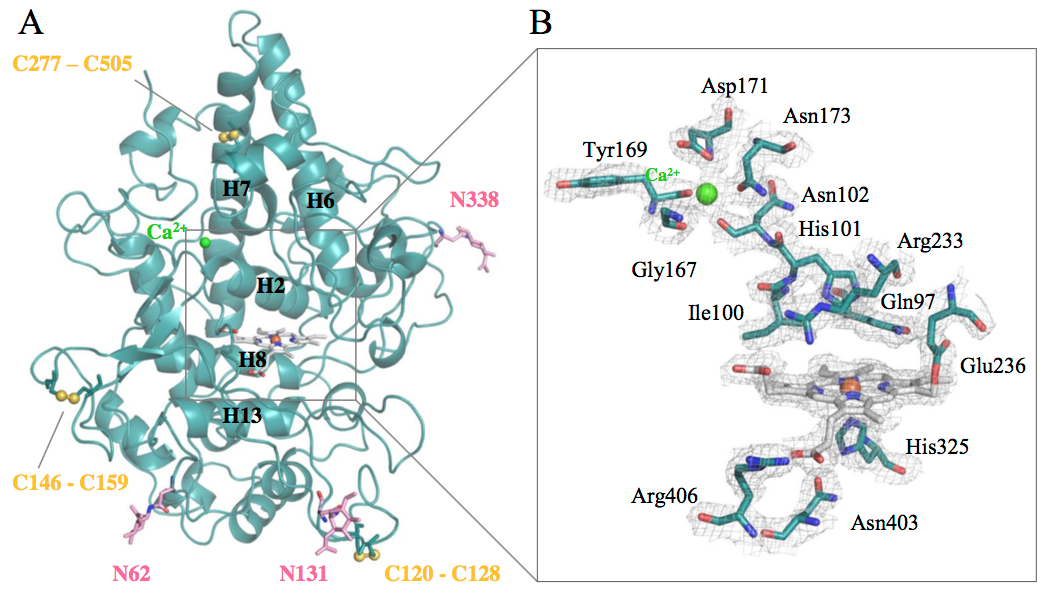
This paper can be read here: A Nicolussi, JD Dunn, G Mlynek, M Bellei, M Zamocky, G Battistuzzi, K Djinović-Carugo, PG Furtmüller, T Soldati, and C Obinger. Secreted Heme Peroxidase from Dictyostelium discoideum: Insights into Catalysis, Structure and Biological Role. JBC 2017, doi: 10.1074/jbc.RA117.000463. http://m.jbc.org/content/early/2017/12/14/jbc.RA117.000463.full.pdf
We have a new PhD in the lab!
Congratulations, Dr. Ana Teresa López-Jiménez!
October 2017
Ethel Bayer Santos is visiting us! 
Ethel, from Universidade de Sao Paulo, will be in our lab for 2 months to study the role of the type VI secretion system of Xanthomonas citri in the interaction with Dictyostelium discoideum. For more info, click here.
September 2017
Welcome to our new PhD student Manon Mottet!

She will study the single-cell dynamics of mycobacterial infection. Click here to learn more about her previous studies.
August 2017
The Dicty 2017 conference, organised this year by Prof. Cosson’s lab and our lab, has been a success!

Exciting science hold in an excellent venue. For more info, click here.
July 2017
We identified new antimycobacterial compounds. Find all the information in our new paper published in PLoS One.
Tuberculosis remains one of the major threats to public health worldwide. Given the prevalence of multi drug resistance (MDR) in Mycobacterium tuberculosis strains, there is a strong need to develop new anti-mycobacterial drugs with modes of action distinct from classical antibiotics. Inhibitors of mycobacterial virulence might target new molecular processes and may represent a potential new therapeutic alternative. In this study, we used a Dictyostelium discoideum host model to assess virulence of Mycobacterium marinum and to identify compounds inhibiting mycobacterial virulence. Among 9995 chemical compounds, we selected 12 inhibitors of mycobacterial virulence that do not inhibit mycobacterial growth in synthetic medium. Further analyses revealed that 8 of them perturbed functions requiring an intact mycobacterial cell wall such as sliding motility, bacterial aggregation or cell wall permeability. Chemical analogs of two compounds were analyzed. Chemical modifications altered concomitantly their effect on sliding motility and on mycobacterial virulence, suggesting that the alteration of the mycobacterial cell wall caused the loss of virulence. We characterized further one of the selected compounds and found that it inhibited the ability of mycobacteria to replicate in infected cells. Together these results identify new antimycobacterial compounds that represent new tools to unravel the molecular mechanisms controlling mycobacterial pathogenicity. The isolation of compounds with anti-virulence activity is the first step towards developing new antibacterial treatments.
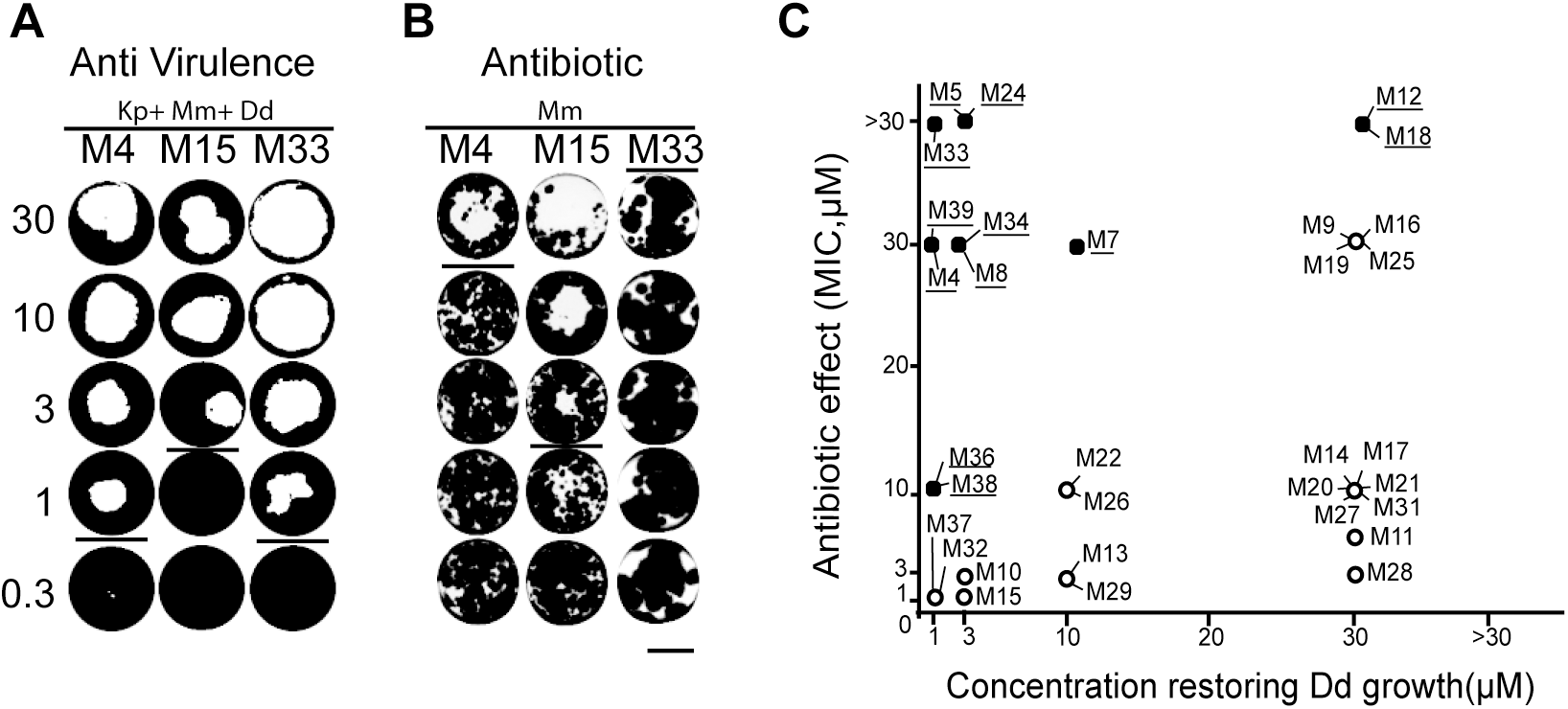
This paper can be read here: Ouertatani-Sakouhi H, Kicka S, Chiriano G, Harrison CF, Hilbi H, Scapozza L, Soldati T, Cosson P. Inhibitors of Mycobacterium marinum virulence identified in a Dictyostelium discoideum host model. PLoS One 2017, 12(7):e0181121. journals.plos.org/plosone/article?id=10.1371/journal.pone.0181121
June 2017
Find here our F1000 recommendation of the article "A Rab20-Dependent Membrane Trafficking Pathway Controls M. tuberculosis Replication by Regulating Phagosome Spaciousness and Integrity", recently published in Cell Host & Microbe by the laboratory of MG Gutierrez.

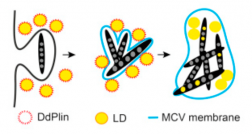
New review by Caroline Barisch: "Breaking fat! How mycobacteria and other intracellular pathogens manipulate host lipid droplets".
This review describes recent evidence about the dual interaction of mycobacteria with host lipid droplets and membrane phospholipids, and integrates them in a broader view of the underlying cellular processes manipulated by various intracellular pathogens to gain access to host lipids.
May 2017
Congratulations to our postdoc Louise Lefrançois for her best poster prize at Mycobactéries 2017 in Université de Versailles-Saint-Quentin-en-Yvelines!

Iuliia Viediernikova, from the Leibniz Institute for Natural Product Research and Infection Biology (Hans-Knöll-Institute, Jena, Germany) got a short-term EMBO fellowship to work with us for two months on the project:
An investigation of the evolutionary origin and defensive role of extracellular traps by social amoeba against filamentous fungi

April 2017
Our new article on how Mycobacterium marinum induces the formation of autophagosomes in its host while repressing the autophagic killing capacity has been published in PLoS Pathogens
One of the cell-autonomous defence pathways against intracellular pathogens is autophagy, an ancestral eukaryotic process surprisingly conserved throughout evolution. Recent studies have highlighted contradictory roles for autophagy during mycobacterial infection. Whereas some studies revealed a role for autophagy to control intracellular bacterial growth, others brought evidence that mycobacteria somehow inhibit autophagic killing. Here, we demonstrate for the first time that Mycobacterium marinum induces both an early autophagic response and its simultaneous repression by blocking the autophagic digestion. This antagonistic manipulation of autophagy is dependent on a functional ESX-1 secretion system, which secretes the membrane-damaging factor ESAT-6, proposed to participate in the perforation of the M. marinum-containing vacuole (MCV). We show here that these membrane damages activate the formation of autophagosomes and their recruitment to the MCV. However, M. marinum also utilizes its ESX-1 secretion system to avoid killing inside autolysosomes by blocking the autophagic flux. In addition, we bring evidence that this manipulation of autophagy is orchestrated via the regulation of TORC1, the major eukaryotic kinase complex controlling nutrient-sensing and cell metabolism.
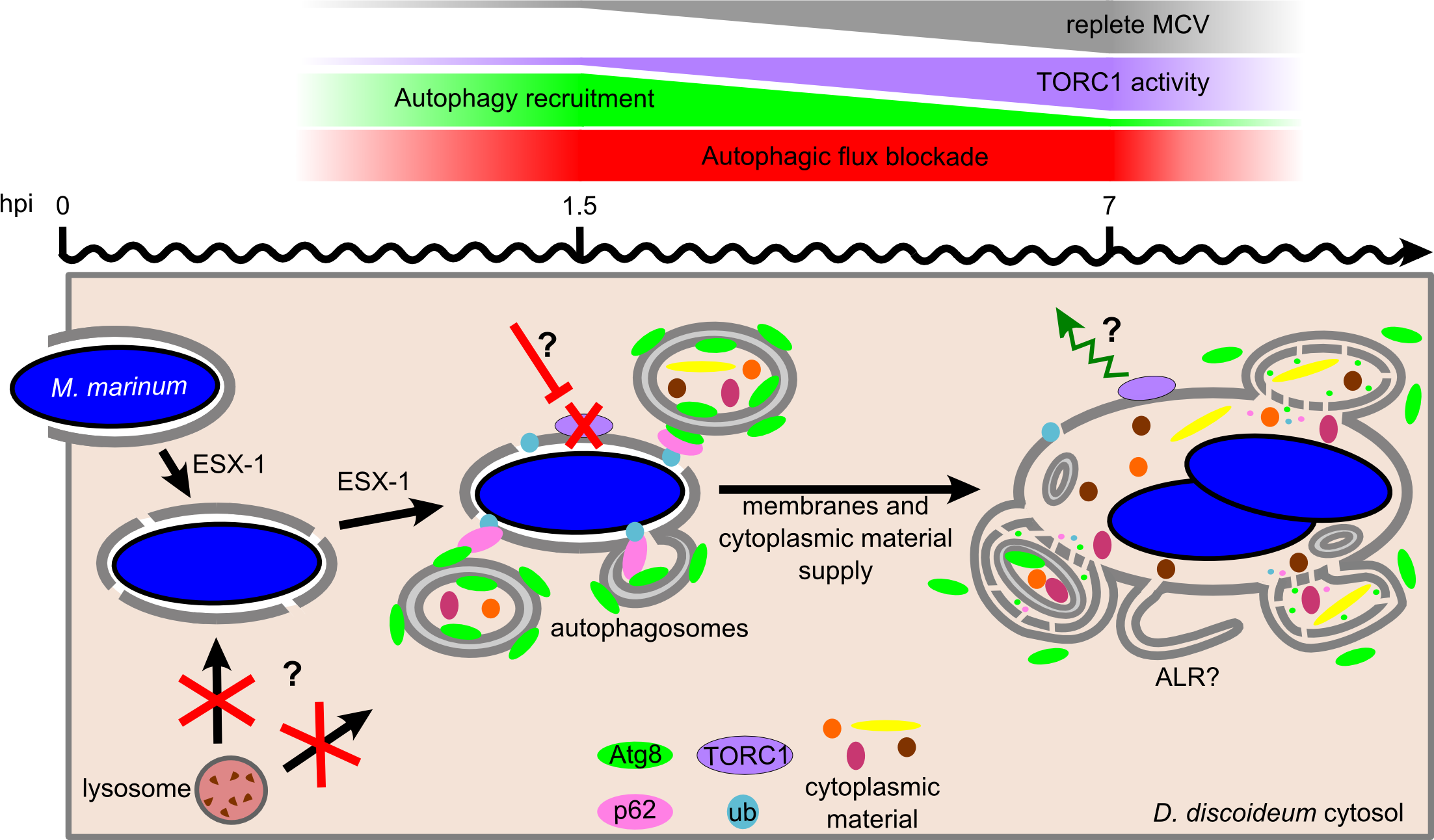
The article can be read here: Cardenal-Muñoz E, Arafah S, López-Jiménez AT, Kicka S, Falaise A, Bach F, et al. (2017) Mycobacterium marinum antagonistically induces an autophagic response while repressing the autophagic flux in a TORC1- and ESX-1-dependent manner. PLoS Pathog 13(4): e1006344. https://doi.org/10.1371/journal.ppat.1006344
January 2017
Mycobacterium marinum not only acquire fatty acids from triacylglycerols stored in host lipid droplets, but also from host phospholipids. Our article has been published in PLoS Pathogens
Mycobacterium tuberculosis (Mtb) survives the human immune defence mechanisms leading to latent tuberculosis in one third of the world population. The ability to persist latently in human macrophages is due to a remarkable physiological change that is accompanied by a slowdown in replication, low metabolism, and phenotypic tolerance to antibiotics. It was proposed that fatty acids released from bacterial intracytosolic lipid inclusions (ILIs), a characteristic of dormant Mtb, serve as carbon source during reactivation from dormancy. In the article “Mycobacterium marinum degrades both triacylglycerols and phospholipids from its Dictyostelium host to synthesise its own triacylglycerols and generate lipid inclusions” we show that the bacteria accumulate ILIs even in a Dictyostelium mutant that is deficient in triacylglycerol synthesis and therefore incapable to build up lipid droplets. In addition, the accumulation of ILIs is not sufficient to induce a dormancy-like phenotype in M. marinum inside its host Dictyostelium. Moreover, we propose an alternative lipid transfer route from the host to the pathogen via degradation and recycling of host phospholipids.
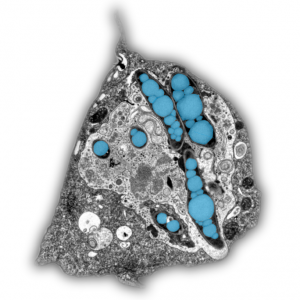
The article can be read here: Barisch C, Soldati T (2017) Mycobacterium marinum Degrades Both Triacylglycerols and Phospholipids from Its Dictyostelium Host to Synthesise Its Own Triacylglycerols and Generate Lipid Inclusions. PLoS Pathog 13(1): e1006095. https://doi.org/10.1371/journal.ppat.1006095
The study has been highlighted in the most various places:
- Docu on Swiss TV, RTS (French)
- Press Release UNIGE (French)
- Swiss newspaper (German)
- Swiss website (German)
- Spanish newspaper (Spanish)
August 2016
Andrea Nicolussi is visiting us! 
Andrea is a PhD student in the Protein Biochemistry Group of Prof. Christian Obinger at the University of Natural Resources and Life Sciences, Vienna. She will be in our lab for 4 months to analyze the function, structure and physiological role of ancestral heme peroxidases.
She tell us more about her PhD project Secreted heme peroxidase from Dictyostelium discoideum – Insights into Catalysis, Structure and Biological Role:
"After producing the metalloprotein recombinantly, we characterized it by various biochemical and biophysical methods, e.g. UV-vis, ECD or EPR spectroscopy, pre-steady state kinetics or differential scanning calorimetry. Solving the X-ray structure of the heme enzyme increased our understanding of the structure-function relationships within the active site. My work on this peroxidase will be complemented by my research stay in the group of Prof. Thierry Soldati in Geneva, where we will investigate the in-vivo role of the peroxidase in the unspecific immune defense of Dictyostelium discoideum.”
MARCH 2016
New lab member!
Welcome to the MSc Lyudmil Raykov, from the Department of Molecular Biology of the University of Geneva.

Our article on the invention of DNA-based extracellular traps by Dictyostelium Sentinel slug cells has been published in Nature Communications
Our innate immune system, made up mainly of phagocytes, protects our body by exterminating bacteria. To do this, it uses two mechanisms. The first kills foreign bodies within the phagocyte itself. The second kills them outside the cell. These two strategies were already known to researchers, but only in humans and other higher animals. In collaboration with our colleagues from the Baylor College of Medicine in Huston (USA) we discovered that the social amoeba Dictyostelium, a unicellular microorganism living in the soils of temperate forests, also uses both these mechanisms, and has done so for over a billion years. Since this amoeba possesses an innate defense system similar to that of humans, while being genetically modifiable, the researchers can therefore carry out experiments on it in order to understand and fight genetic diseases of the immune system. This discovery can be read in the journal Nature Communications.
To defend themselves, our immune cells have two mechanisms. The first, called phagocytosis, kills bacteria within the phagocytic cell itself. The cell envelops the foreign body and exterminates it specifically by using reactive oxygen species (ozone, hydrogen peroxide, bleach), generated thanks to the enzyme NOX2. However, when the invader is too large to be taken up, cells use a second defense mechanism which consists of expelling their genetic material, that is to say their DNA. This DNA transforms into sticky and poisoned nets called «neutrophil extracellular traps» (NETs). These DNA nets then capture bacteria outside of the cell and kill them.
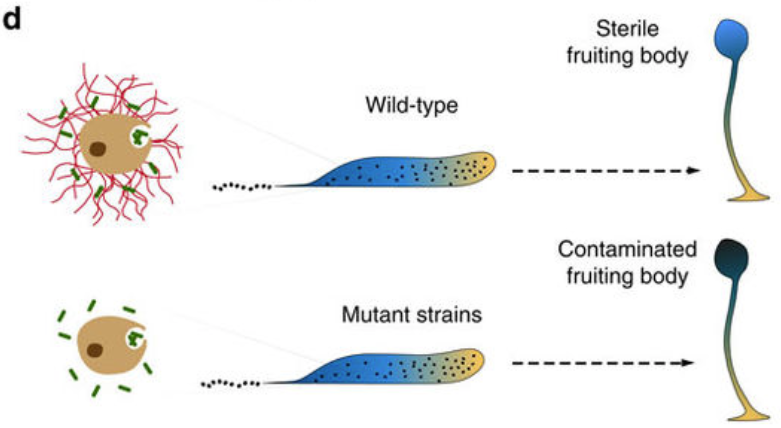
The article can be read here: Zhang, X., Zhuchenko, O., Kuspa, A., and Soldati, T. (2016) Social amoebae trap and kill bacteria by casting DNA nets Nature Communications 1;7:10938. doi: 10.1038/ncomms10938.
The study has been highlighted in the most various places:
- Université de Genève, Service de Communication (French)
- EurekAlert, The Global Source for Sciences News
- Sveriges Radio (Swedish)
- Quo (Spanish)
January 2016
New lab member!

Welcome to the PhD Louise Lefrancois, from the Research institue of the McGill University Health Centre (Montreal, Canada).
February 2015
We have a new PhD in the lab!
Congratulations, Dr. Valentin Trofimov!

December 2013
We have a new PhD in the lab!
Congratulations, Dr. Xuezhi Zhang!
November 2013
We have been awarded a COST grant from the Swiss State Secretariat for Education, Research and Innovation
This project aims to study cellular and molecular functions of ROS and NOX in a social amoeba model system.
During this project supported by the COST / SEFRI, we propose to use our simple and powerful experimental model system Dictyostelium infected with Mycobacterium marinum to dissect functions of ROS/NOX in host pathogen interactions, phagosomal bactericidal activities, including metal poisoning, but also in chemotaxis and morphogenesis.
We have become a member of iGE3

The mission of the Institute of Genetics and Genomics of Geneva (iGE3) is to promote internationally competitive biomedical research and high quality teaching by using primarily genetic and genomic scientific analysis.
"iGE3 is a dynamic structure of the University of Geneva to promote research and teaching related to the human and other genomes. Our ambition is to understand life in the light of the structure, variation and function of genomes, and to contribute to the promotion of health based on the studies of the various human genomes." The major aims of the Soldati group are to understand the integration, the cooperation of signalling, cytoskeleton and membrane trafficking in phagocytosis and its relevance to host-pathogen interactions.
We have been awarded a SystemsX grant

The HostPathX project aims to study the host-pathogen interface, using modelling and chemical genetics perturbation of the phagocyte – mycobacteria interface.
This collaborative grant is a partnership with the labs of:
- Pierre Cosson at the Department of Cell Physiology and Metabolism of the university of Geneva
- Hubert Hilbi at Institute of Medical Microbiology of the University of Zurich
- Marco Pagni at the Swiss Institute of Bioinformatic in Lausanne
- Heinz Koeppel at the BISON Group | D-ITET | ETH Zurich
During this 4 year SystemsX.ch RTD project, we propose to identify the mode of action of anti-infective compounds by innovative methodologies that probe the perturbations of the host-pathogen interaction landscape. We will exploit the technological developments in high throughput RNA-sequencing to determine transcriptional signatures triggered by compounds that decrease bacterial pathogenicity. Notably, we will analyze the transcriptional response of host cells to infection with bacteria, and its perturbation by anti-infective compounds. Through developing novel computational methods and leveraging state-of-the art network analysis and modelling tools, we will predict a putative mode of action for each compound, and test the hypotheses directly using appropriate biological assays. To this end, we will exploit the genetic tractability of both Dictyostelium and Mycobacterium marinum.
October 2013
We have a new PhD in the lab!
Congratulations, Dr. Aurélie Guého!
May 2013
The Soldati Group had a productive retreat
The Soldati lab has come back from their scientific retreat in Hotel Bellevue Le Rocheray, in the Vallée de Joux. It took place on the 30th and 31th May.
DECember 2012
We have become a member of the BMBS COST Action BM1203
The EU-ROS COST Action aims to study the functions of ROS and NOX at all levels, from bench to bed side.
Unravelling the fine balance between Reactive Oxygen Species (ROS) acting as a friend or a foe is fundamental to understand aerobic life. To advance this important area of biology and medicine, highly synergistic approaches combining diverse and scattered disciplines are needed. For this, COST provides an ideal framework. EU-ROS brings together multi-disciplinary experts to enhance the competitiveness of European research. Collectively, EU-ROS will overcome the fragmentation of European R&D on oxygen/ROS research while its translational components will contribute to European societies’ economic growth and wellbeing.

 Have a look here!
Have a look here!