Techniques used in the Nouspikel lab
We use a whole range of classical laboratory techniques:
- Cell biology: cell culture, transfection
- Molecular biology: DNA/RNA purification, cloning, PCR, siRNA.
- Proteomics: Western blots, 2D gels, protein purification.
But we also use a few specialized techniques to study DNA repair, which
are briefly described below.
Global genome repair (GGR) of UV-induced
lesions
GGR of benzopyrene chemical adducts
Gene specific repair of UV-induced lesions (TCR
and DAR)
In-vitro assay for cisplatin crosslinks
Click on any picture to enlarge it.
Global repair of UV-induced lesions
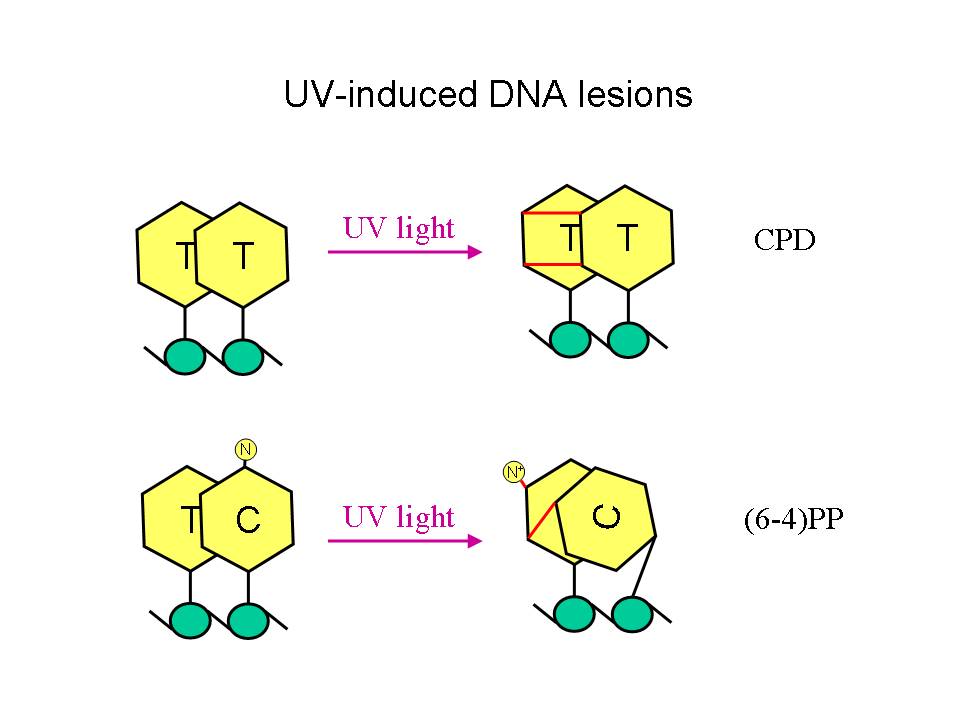 UV
light causes two types of DNA lesions: cyclobutane-pyrimidine dimers
(a.k.a. CPDs)
and (6-4)pyrimidine-pyrimidone photoproducts (a.k.a (6-4)PPs). Both are
characterized by the formation of illegitimate bonds between two
pyrimidines adjacent on the same DNA strand.
UV
light causes two types of DNA lesions: cyclobutane-pyrimidine dimers
(a.k.a. CPDs)
and (6-4)pyrimidine-pyrimidone photoproducts (a.k.a (6-4)PPs). Both are
characterized by the formation of illegitimate bonds between two
pyrimidines adjacent on the same DNA strand.
We can generate these lesions very easily by briefly irradiating cells
with a germicidal lamp. The big advantage of this technique over
chemical agents is that we can control very precisely the amount of
damage and the moment when it is introduced.
We can quickly and easily measure the amount of each lesion in bulk
genomic DNA thanks to antibodies produced by Dr Toshio Mori, which
specifically recognize CPDs or (6-4)PPs. DNA is simply blotted on a
membrane, and the amount of lesions is measured in a "western blot"
type of experiment.
Alternatively, we can use an assay relying on the property of the phage
enzyme T4 endonuclease V (TEV) to nick the damaged strand in proximity
of a CPD. We simply digest DNA with TEV, denature it to separate the
strands, and measure the size of the fragments (for instance with
sucrose gradients). The more lesions, the smaller the fragments.
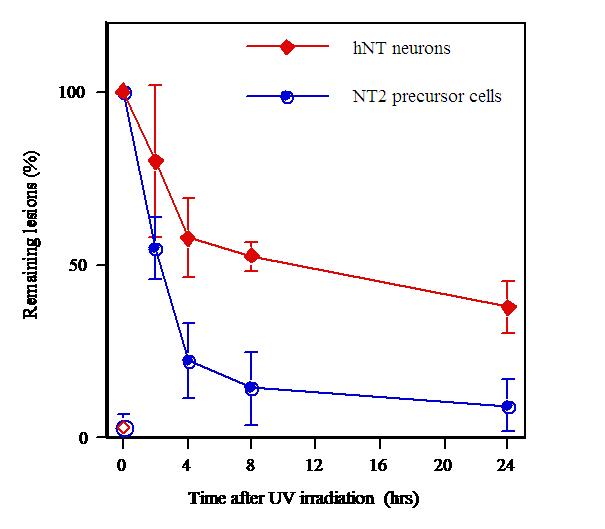
Example
Removal of (6-4)PPs from hNT neurons (in red), compared with their
precursor cells (NT2, in blue), measured with the antibody assay. Click
to enlarge.
For more details:
Terminally differentiated human
neurons repair transcribed genes but
display attenuated global DNA repair Nouspikel,T.
& Hanawalt,P.C. Mol.
Cel. Biol.
20:1562-1570
(2000) (download pdf file
138 K)
Global repair of BPDE adducts
Benzopyrenes are chemicals present in cigarette smoke, which forms
benzo[a]pyrene diol-epoxide (BPDE) adducts on purines. To reveal the
lesions, we use an assay developed by Dr Dave Philips, in which DNA is
digested to the nucleotide level with nucleases. The presence of
a BPDE adduct inhibits the nuclease and results in the formation of
di-nucleotides, which can be post-labeled with 32P and revealed by
2-dimensional thin layer chromatography.
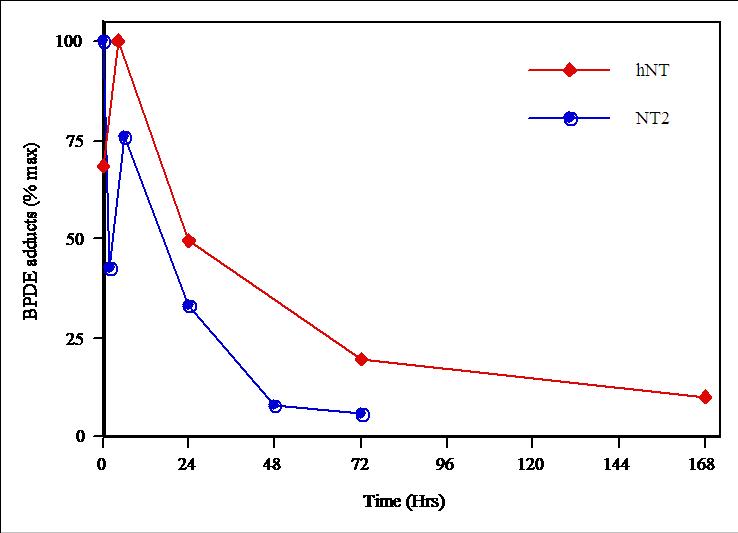
Example
Repair of BPDE adducts in NT2 precursors (blue) and hNT neurons (red).
The initial dose of BPDE is comparable to what is found in the blood of
a heavy smoker...
For more details:
p53-dependent global genomic repair of
benzo[a]pyrene-7,8-diol-9,10-epoxide adducts in human cells. Lloyd,D.R.
& Hanawalt, P.C. Cancer Res. 60:517-521 (2000)
TCR and DAR assay
Transcription-coupled repair (TCR) and differentiation-associated
repair (DAR) are much more difficult to measure, because we need to
work at the level of a single gene. These experiments are
time-consuming (about 2 weeks) and require a lot of DNA (50 ug per
condition). The assay, developed in the 80s in the Hanawalt lab,
is basically a Southern blot with a couple of twists.
First of all, in addition to the required restriction enzyme, DNA is
also treated with TEV, which will chop any restriction fragment
containing a CPD. Thus, on the final autoradiogram, only those
fragments that did not contain any CPD will appear as a nice band. By
quantifying this band, and by knowing the initial amount of fragments
(which we get from a mock-digested control), we can deduce the amount
of CPDs by using the Poisson expression:
Amount of lesions = -ln(intact fragments/total fragments)
The other trick is to use RNA probes so as to reveal one strand at a
time. This allows us to visualize differences in repair between the
transcribed strand and the non-transcribed strand. Typically, TCR will
cause a strand bias in an active gene, with the TS being repaired
faster than the NTS. DAR is defined as the proficient repair of the NTS
in cells that have little or no global repair of CPDs.
Example
TCR and DAR in the cMyc gene in HL60
cells.
For more details
Determination of damage and repair in
specific DNA sequences. Spivak,G.
& Hanawalt,P.C. Methods 7:147-161 (1995)
In-vitro assay for cisplatin
crosslinks

This assay was developed by the laboratory of Rick Wood. In its most
basic version, a plasmid is build, which contains an unique cisplatin
crosslink in the vicinity of a 32P label. The plasmid is then incubated
with whole cell extract, prepared according to Manley et al. If NER
occurs, it will excise a short (around 29 nt) oligonucleotide
containing both the lesion and the label. These excision fragments are
very specific and can be revealed on a sequencing gel.
Repair-deficient extracts can then be complemented with various protein
fractions, to try and identify the cause of the deficiency.
For more details
Mammalian DNA nucleotide excision
repair reconstituted with purified
protein components. Aboussekhra,A.
et al. Cell 80:859-868 (1995)
More browsing
Research interests
Thierry Nouspikel's CV
 UV
light causes two types of DNA lesions: cyclobutane-pyrimidine dimers
(a.k.a. CPDs)
and (6-4)pyrimidine-pyrimidone photoproducts (a.k.a (6-4)PPs). Both are
characterized by the formation of illegitimate bonds between two
pyrimidines adjacent on the same DNA strand.
UV
light causes two types of DNA lesions: cyclobutane-pyrimidine dimers
(a.k.a. CPDs)
and (6-4)pyrimidine-pyrimidone photoproducts (a.k.a (6-4)PPs). Both are
characterized by the formation of illegitimate bonds between two
pyrimidines adjacent on the same DNA strand.

