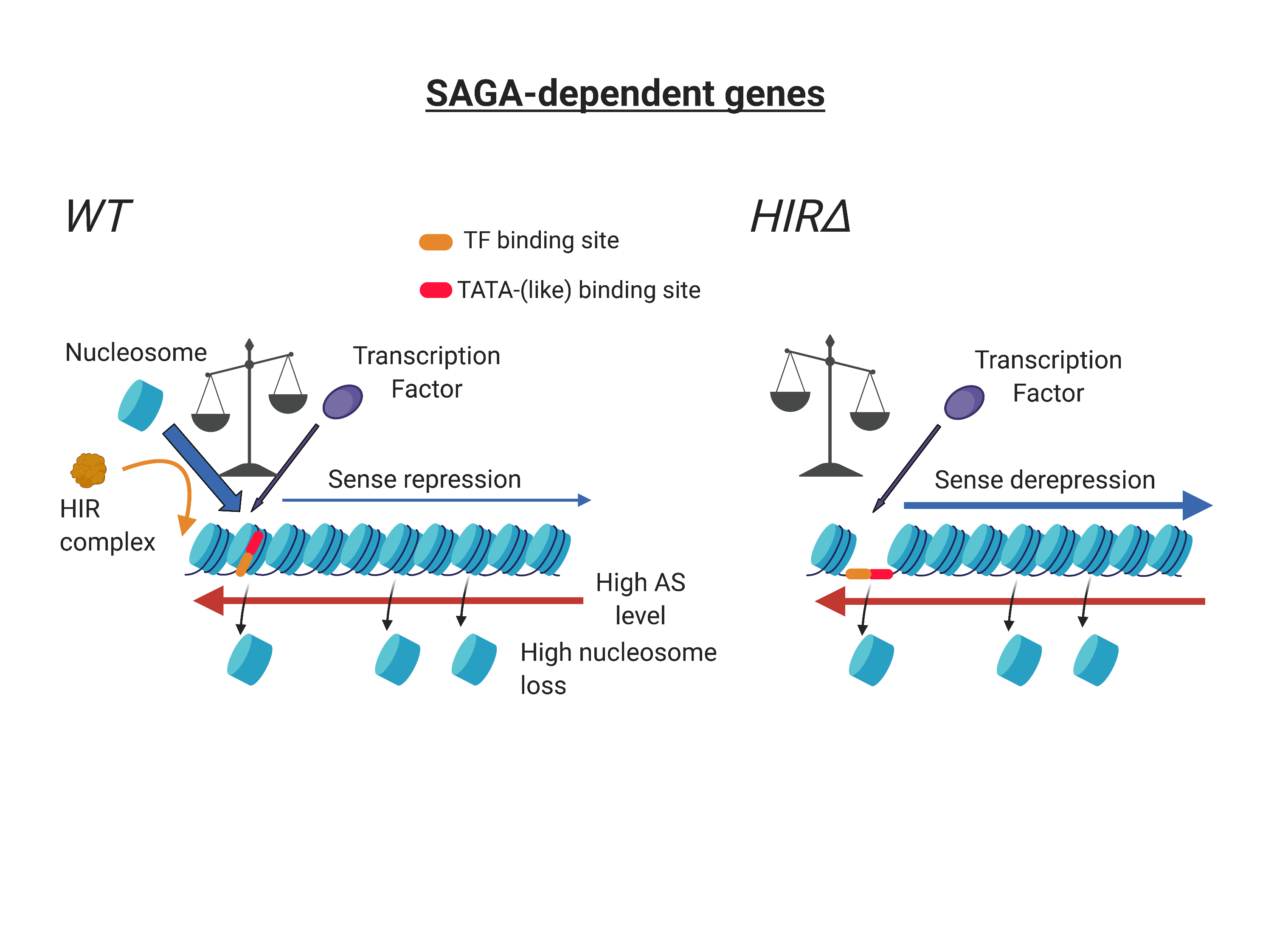
Antisense-mediated repression of SAGA-dependent genes involves the HIR histone chaperone
While the spotlight has been for a long time on coding transcription, it turns out that noncoding transcription is largely predominant in a eukaryotic cell. The pervasiveness of non-coding transcription might have functional consequences: many non-coding transcripts overlap with promoter regions of coding genes. This might lead to the repression of the corresponding coding gene via a mechanism of transcription interference. What are the coding genes affected by transcription interference?
The team of Françoise Stutz from the Department of Molecular and Cellular Biology proposes that the targeted coding genes belong to a specific class called SAGA-dependent genes. These genes, representing 15% of coding genes, show a closed yet very dynamic chromatin at their promoter, and are more subject to expression heterogeneity in the cell population. This work reveals the importance of non-coding transcription in proper gene regulation.
This study has been performed using the baker’s yeast as a model organism. However, considering the conservation of the involved factors, it is likely that the regulation of many human coding genes may depend on the proposed mechanism.
This article was published in the journal Nucleic Acids Research on April the 27th.