[895] Bacterial glyco-biology and chemical-genetics
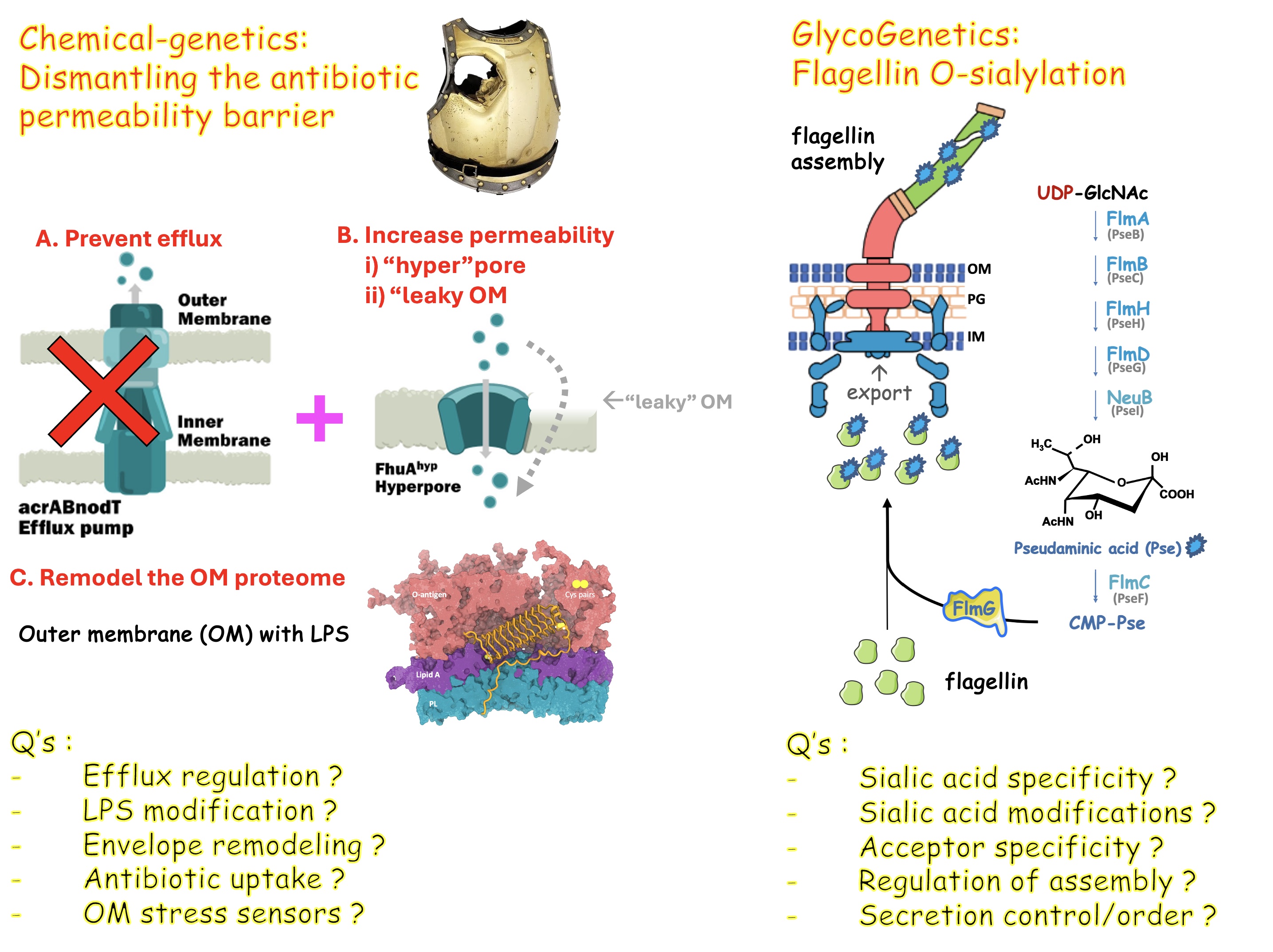
We use cutting-edge glyco-genetic and chemical-genetic approaches to unravel two fundamental questions, addressed with (Gram-negative) bacteria as models.
Why and how do bacteria decorate their surface structures such as the flagellum with sialic acids, compared to animals? How is sialic acid covalently and conjugated to specific proteins or polysaccharide structures? How are the sialic acids derivatized and how is this synthesis regulated?
Another research focus is on the unusual role of the bacterial outer membrane (OM) in counteracting antibiotic action, crippling antibiotic entry and by promoting their efflux. How is the OM integrity ensured, how is it chemically modified and how is efflux regulated? Can multi-drug resistance (MDR) of certain bacteria can be overcome? Can we modify antibiotics to overcome bacterial MDR?
