[1036] RNA metabolism and bacterial pathogenesis
→ The bacterial opportunistic pathogen Pseudomonas aeruginosa colonizes a wide range of environments and hosts and it causes multiple types of life-threatening infections in humans. Contrary to host-specialized pathogens, P. aeruginosa builds its success as a pathogen on adaptability and it possesses one of the largest and most intricate regulatory systems among bacteria to sense and respond to multiple environmental cues.
Overall, our research aims at understanding of broad questions on bacteria environmental adaptation and pathogenesis focusing on P. aeruginosa and the control of gene expression exerted by RNA-binding proteins (RBPs). RBPs are considered the fastest and the most flexible among all types of gene regulators, but they are the least characterized in bacteria.
By binding to RNA molecules, RBPs can affect RNA degradation and/or translation of mRNAs. In our work, we characterize the action of novel RBPs by using two approaches:
A) Molecular and biochemistry techniques to identify their mode of action ;
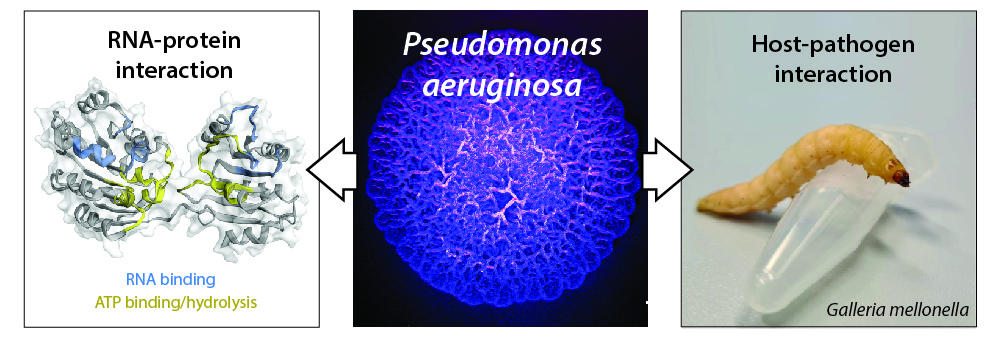
B) Microbiology assay and in vivo infection assays using larvae of Galleria mellonella as host organism to study of the impact of an RBP on bacterial virulence and host-pathogen interaction.
Interestingly, some RBPs have been proposed as drug target, being widespread in bacteria but with no homologs in humans. Our work will elucidate this possibility and foster the development of new antimicrobial drugs targeting RBPs.
Figure legend: (mid panel) surface of a Pseudomonas aeruginosa colony biofilm (stereomicroscope, artificial colour); (left panel) model of the RhlE2 RNA helicase structure and putative regions involved in RNA binding and ATP binding/hydrolysis (Hausmann et al. 2021); (right panel) photo of a Galleria mellonella larva, 3R-compatible organism to study host-pathogen interactions.