Biophysics of the Gradient
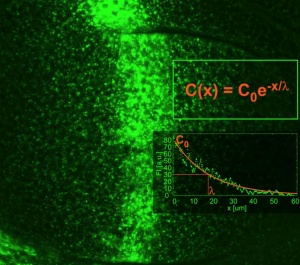
Cells in a tissue know about their position by reading the gradient of concentration of morphogens. Morphogens are transcribed in clusters of cells from developing tissues and from there the morphogen is secreted and diffuse to form a gradient of concentration as the molecules are diluted away from the source.
The conceptof morphogen gradient is deeply quantitative: cells respond with signaling above a threshold concentration value. We have been interested on what controls the formation of the gradient in physical and cell biological terms. We focused in a particular morphogen that operates during Drosophila wing development: the TGF-beta type morphogen Dpp.
We understand it in physical terms: it depends on the diffusion coefficient, the rate of degradation, the rate of production in the source. At the cell biological level, we found that endocytosis of the ligand is important for its movement. Perhaps the morphogens are moving through the cells by endocytosis and recycling, rather than around the cells by free diffusion.
Biophysics of tissue growth and morphogens
We have then studied in a highly quantitative way the behaviour of the gradient as the developing tissue grows. We think that the cells can compute the increase in signaling over time and use this information to control growth.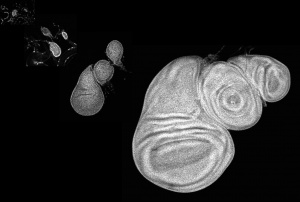
Also, the gradient changes its properties during growth: it flattens because the Dpp degradation rate decreases during development and this contributes to keep cells increasing their signaling level over time. We want to understand the physics and molecular biology of this flattening phenomenon. Which factor controlling lysosomal degradation of the morphogen can compute the size of the tissue to fine tune the Dpp degradation rate?
We also want to understand what is the noise of the system, how it fluctuates, because this must affect the computation of signaling over time.
Of course is also important to figure out how cells can measure signaling over time: "how do cells measure time derivatives?". To study this we are engineering signaling biosensors that allow us to study in real time the signaling levels in individual cells as the tissue grows.
Our model system to study this has been the Drosophila fly wing. We are approaching with the same quantitative, physics strategy the morphogen-dependent growth of the pectoral fin of Zebrafish.
Sara endosomes
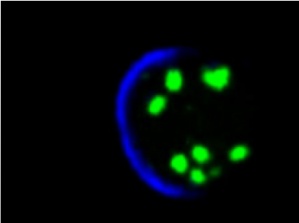
We speculate that, to measure signaling over time, cells need a memory of the signaling level. Some of us think that this memory reside in the endosomal pool of molecules. We study a particular endosome: the Sara endosome.
We have studied these Sara endosomes in the past and found that they are dispatched symmetrically across mitosis by using the mitotic spindle. This ensures that, in a morphogen gradient situation, daughter cells after mitosis "remember" the signaling levels of the mother.
We then studied the Sara endosomes during asymmetric division. In the sensory organ precursors and in stem cells. We focused on neural stem cells and intestinal stem cells. We found that in these situations the Sara endosomes contain the key signaling molecules there, Notch and Delta. In this set up, the Sara endosomes also go to the spindle, but this time they move asymetrically into only one of the daughter cells. And this is essential to bias the signaling event. Also during the mitosis of the neural precursors of a vertebrate, the Zebrafish. In Zebrafish, we are also looking at the Anthrax receptors, because they turn out to control the polarized mitosis of cells during early embryogenesis, a process which is behind the convergence-extension movements that shape the fish embryo.
We are now studying the biophysics and the molecular machineries behind the directed motility of the Sara endosomes. We developed many assays for this, among other things the generation of magnetic endosomes whose motility we can direct at our will using magnets.
Lipidomics in fish and flies
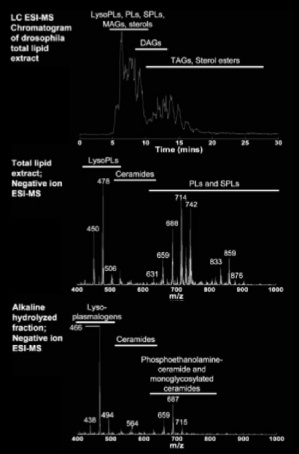
We think that the lipid composition of these compartments, which is largely unknown, form part of the molecular machinery that endows this particular behaviour. In flies and fish, the lipids are largely unexplored. We initiated a lipidomics project to understand the role of lipids during development. We do this in collaboration with the Wenk lab in Singapore in the context of the LipidX consortium. With them we are learning to quantitate and characterize the highly rich lipid repertoire during the development of flies and fish.
