Detecting DNA knots to inhibit pathogens
Scientists from the University of Geneva (UNIGE) Faculty of Medicine have elucidated how the SMC5/6 protein complex detects extrachromosomal DNA, a property that enables it to fight viral infections. This discovery could have implications beyond the antiviral response. Indeed, many cancer cells harbour extrachromosomal DNA, opening new avenues for research in oncology. These results can be read in the journal Nature Communications.
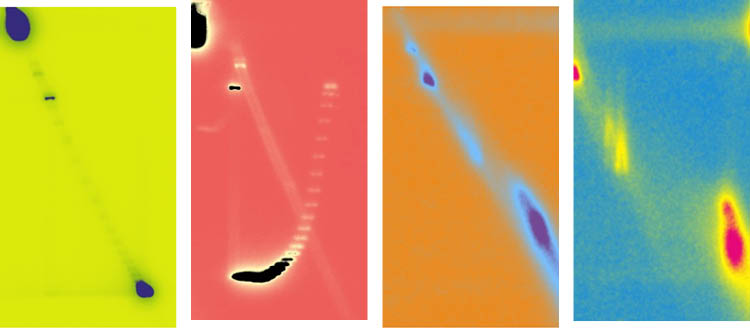
Aurélie Diman, UNIGE. Pop Art’ autoradiographic image illustrating the migration of circular extrachromosomal DNAs of opposite topology during two-dimensional agarose gel electrophoresis (2D). Image modified with https://www.bloggif.com/pop-art.
The SMC5/6 complex is vital for maintaining genome stability, playing a crucial role in chromosome organization and DNA repair. In 2016, Professor Michel Strubin's team from the Department of Microbiology and Molecular Medicine at the UNIGE Faculty of Medicine discovered that this complex is a key player in antiviral defense, especially against the hepatitis B virus. Since then, SMC5/6 has demonstrated effectiveness against several other human pathogenic viruses, expanding its scope in the fight against infection.
Aurélie Diman, a post-doctoral fellow in the Department of Microbiology and Molecular Medicine, describes the discovery made with her colleagues: “Since 2016, we knew that the SMC5/6 complex can bind to the hepatitis B genome and any DNA artificially introduced into cells, as long as this DNA remains extrachromosomal, i.e. outside the host cell's chromosomes. However, once this DNA integrates into the chromosomes, the binding is lost. This suggests that the SMC5/6 complex can detect a specific feature of extrachromosomal DNA. It is this specific signature that we have identified.”
Targeting extrachromosomal DNA
To function, the SMC5/6 complex binds to extrachromosomal DNA and then transports this "foreign" genetic material to a subcompartment of the cell nucleus, where it blocks its expression. During a viral infection, this process prevents the virus from replicating, positioning SMC5/6 as both a detector and an inhibitor. "The fundamental question was to understand how the SMC5/6 complex identifies which DNA fragments it needs to target", explains Aurélie Diman.
Detecting DNA knots
Using both viral and non-viral extrachromosomal DNA, scientists have demonstrated that the SMC5/6 complex in our cells preferentially binds to circular extrachromosomal DNA rather than linear DNA, and shows a particular affinity for highly expressed DNA. When DNA is expressed, it can twist in various directions, forming specific structures such as supercoils or knots. These unique structures act as markers, enabling SMC5/6 to identify its targets.
The scientists were able to reveal this using a somewhat forgotten technique: two-dimensional agarose gel electrophoresis. This method allows for the identification of variations in DNA molecule shape or its topology. The technique involves two successive migration steps: the first separates DNA molecules based on their size, while the second, performed perpendicularly, distinguishes them based on their topology. "Despite technological advancements and the development of new methods, this technique remains the gold standard for determining the direction of DNA twisting," notes Aurélie Diman.
Are cancer cells also affected?
The implications of this discovery extend far beyond the field of virology. Circular extrachromosomal DNAs, which strongly expresses genes that promote tumor growth, are found in a significant proportion of cancers. " Our next goal is to deepen our understanding of the molecular mechanisms that regulate the activity of the SMC5/6 complex and to determine how extrachromosomal DNA in cancer cells evades its control " conclude the authors.
9 Sept 2024