[1037] Function and mechanism of germline small non-coding RNAs in sperm and embryos
The lab
We are interested in different aspects of male reproduction, including spermatogenesis, sperm function, and intergenerational transmission of paternal information. Specifically, we are passionate about understanding how the interplay between germline small RNA, mRNA regulation, and RNA-binding proteins leads to robust reproductive fitness. Several lab projects use genetically modified mice, imaging, biochemistry, sequencing, and bioinformatic analysis to answer interesting questions about the genetic and molecular mechanisms of reproduction. We foster a collaborative and inclusive work environment and are part of the iGE3, NCCR RNA & Disease, and ANDRONET Cost Action networks.
Male reproduction
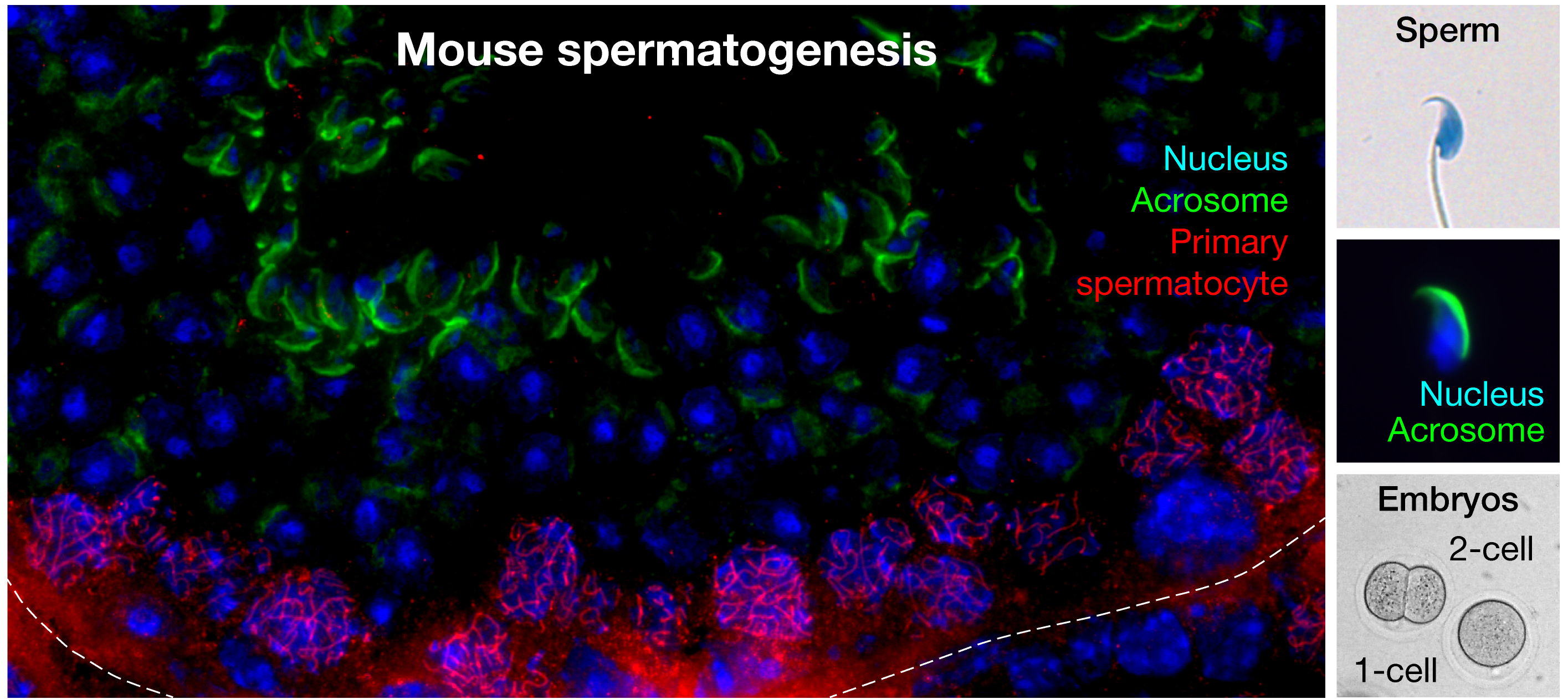
We study the mechanisms of male reproduction from multiple angles: we explore how sperm RNA is selected during spermatogenesis, how RNA-binding proteins may participate in sperm RNA selection, and how male mouse-specific germline small RNAs—piRNAs—contribute to paternal regulation of embryonic gene expression. By understanding how spermatogenesis is regulated and how sperm contributes to embryo viability, our work could inform new recommendations to improve male fertility and medically assisted reproduction outcomes.
PIWI-interacting RNA (piRNA)
We are interested in the function and mechanisms of piRNAs—a class of germline small RNAs that protect animal fertility by guiding PIWI-clade Argonaute proteins to silence transposons or regulate mRNAs. In adult mouse testes, a subclass called pachytene piRNAs is produced in vast numbers. These piRNAs regulate mRNAs critical for spermatogenesis and sperm function through partial guide-target complementarity. While their role in male fertility in both mice and humans is well established, the biological significance of their remarkable abundance and sequence diversity remains an open question. We investigate the possibility that pachytene piRNAs serve a paternal role after fertilization, which may partly explain their high abundance and diversity.
