Chromatin regulation in brain development and disease
Our goal is to understand what drives brain cell diversity during development. We focus on the role of epigenetic regulators that control chromatin accessibility and gene expression. The lab uses epigenome editing technologies, mouse stem cell cultures, in vivo models and state-of-the-art genomic techniques to study mechanisms of chromatin regulation in the brain. Our aim is to gain novel insights into neurodevelopmental disorders like autism, which are frequently associated with mutations in epigenetic regulators.
Brain development. Characterizing the diversity and function of brain cell types is essential to our understanding of complex brain functions and neurodevelopmental disease. What remains poorly understood is what establishes the gene expression networks that determine a brain cell’s identity and how misregulation of this epigenetic process causes disease. We seek to unravel the molecular mechanisms that allow cell type-specific chromatin remodeling complexes to regulate chromatin accessibility at genes that promote distinct cell fates.

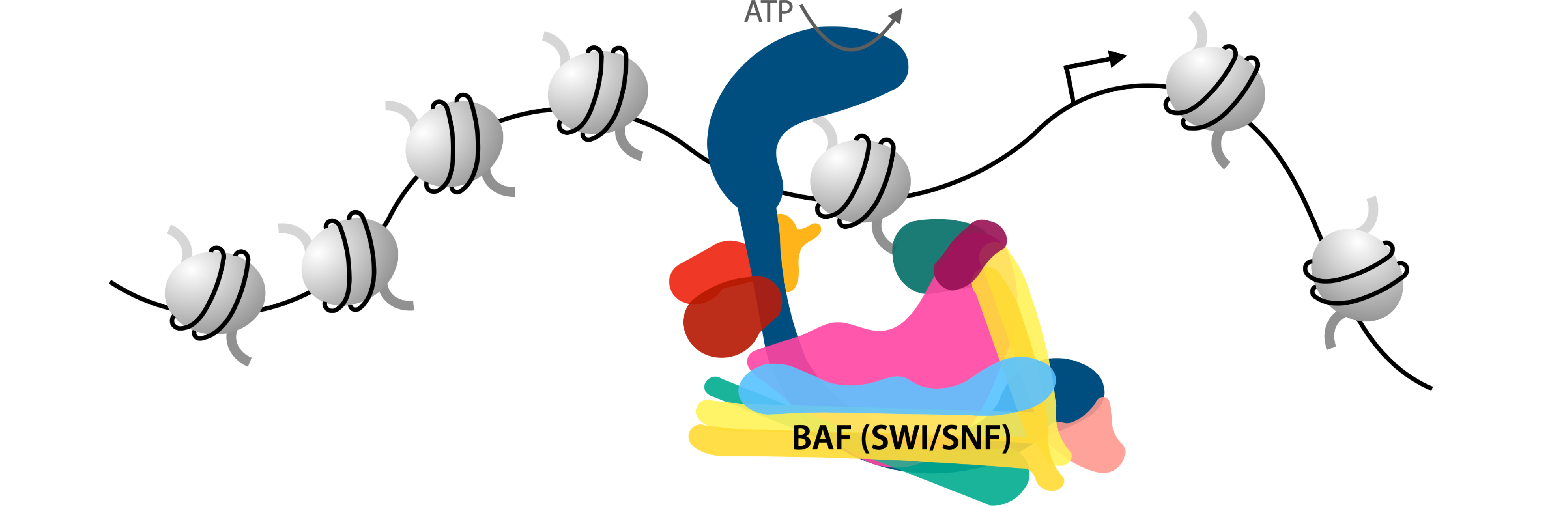
Chromatin remodeling. Chromatin remodelers generate chromatin accessibility through ATP-dependent nucleosome eviction and repositioning. These multi-subunit complexes can be assembled into cell type-specific forms to regulate chromatin in a cell type-specific manner. A key driver of brain cell fate is the BAF (or mSWI/SNF) chromatin remodelling complex. There are hundreds of alleles of BAF complex subunits that have been linked to human neurodevelopmental disorders. We investigate how BAF complexes regulate cell type-specific gene expression networks during brain development. Our goal is to shed new light on the general epigenetic principles that control brain cell fate as well as reveal the molecular mechanisms that characterize neurodevelopmental disorders.
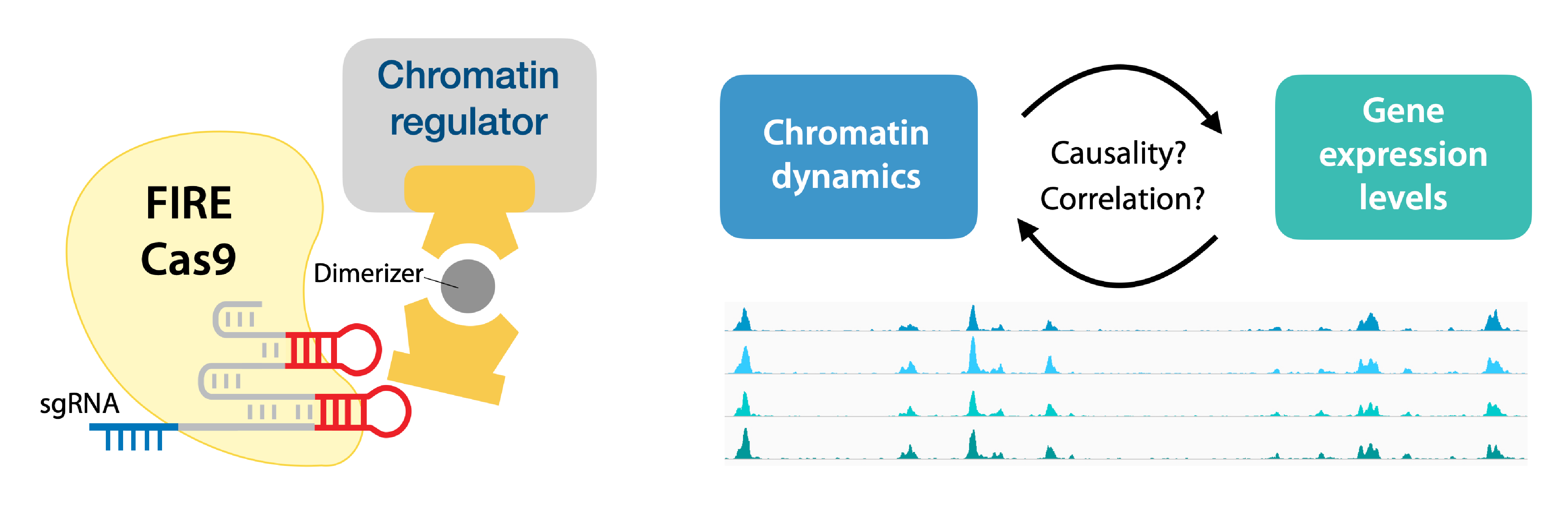
Epigenome editing. Understanding the causal link between epigenetic marks and gene regulation remains a central question in chromatin biology. To edit the epigenome we developed the FIRE-Cas9 system for rapid and reversible recruitment of endogenous chromatin regulators to specific genomic loci. This strategy provides precise kinetic information to model epigenetic memory and plasticity in living cells. It is broadly applicable to mechanistic studies of endogenous multi-subunit chromatin regulator complexes.